|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.24 |
Binding ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2480 |
0.26 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.33 |
Binding ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
280 |
0.31 |
Functional ≤ 10μM
|
|
Z80135-1-O |
F9 (cluster #1 Of 1), Other |
Other |
33 |
0.35 |
Functional ≤ 10μM
|
|
Z80140-1-O |
GBM (cluster #1 Of 1), Other |
Other |
550 |
0.29 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
490 |
0.29 |
Functional ≤ 10μM
|
|
Z80250-1-O |
Me665/2/21 (cluster #1 Of 1), Other |
Other |
420 |
0.30 |
Functional ≤ 10μM
|
|
Z80306-1-O |
NB-4 (Promyelocytic Leukemia Cells) (cluster #1 Of 1), Other |
Other |
400 |
0.30 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
130 |
0.32 |
Functional ≤ 10μM |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
430 |
0.30 |
Functional ≤ 10μM
|
|
Z80449-1-O |
SAOS-2 (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
670 |
0.29 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
440 |
0.30 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
510 |
0.29 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
680 |
0.29 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
430 |
0.30 |
Functional ≤ 10μM
|
|
Z81065-1-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
350 |
0.30 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
320 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.63 |
13.65 |
-51.5 |
1 |
3 |
-1 |
60 |
397.494 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.27 |
Binding ≤ 10μM |
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
34 |
0.34 |
Binding ≤ 10μM |
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.31 |
Binding ≤ 10μM |
|
Z80135-1-O |
F9 (cluster #1 Of 1), Other |
Other |
37 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.69 |
15.7 |
-51.53 |
0 |
3 |
-1 |
49 |
411.521 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
5',5',8',8'-TETRAMETHYL-5',6',7',8'-TETRAHYDRO-2,2'-BINAPHTHALENE-6-CARBOXYLICACID
5',5',8',8'-TETRAMETHYL-5',6',7'…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
580 |
0.32 |
Binding ≤ 10μM |
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
52 |
0.38 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
68 |
0.37 |
Binding ≤ 10μM
|
|
Z102258-1-O |
Trachea (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80135-1-O |
F9 (cluster #1 Of 1), Other |
Other |
15 |
0.41 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.43 |
3.07 |
-52.12 |
0 |
2 |
-1 |
40 |
357.473 |
2 |
↓
|
|
|
|
Analogs
-
2568233
-
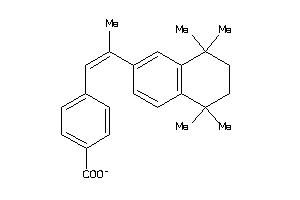
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
85 |
0.38 |
Binding ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.45 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Functional ≤ 10μM |
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Functional ≤ 10μM |
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM |
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80135-1-O |
F9 (cluster #1 Of 1), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.92 |
14.62 |
-49 |
0 |
2 |
-1 |
40 |
347.478 |
3 |
↓
|
|
|
|
Analogs
-
12484958
-
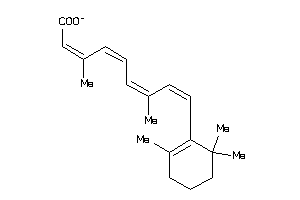
-
22066351
-
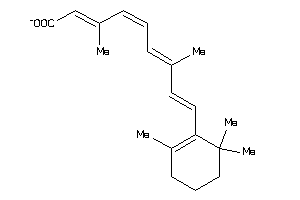
-
33943508
-
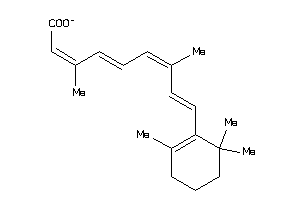
-
4430042
-
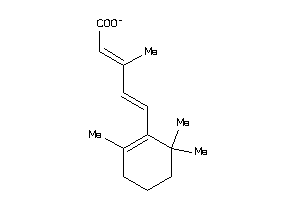
-
9212654
-
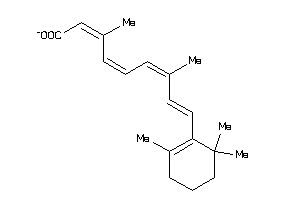
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RABP1-1-E |
Cellular Retinoic Acid-binding Protein I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
RABP2-1-E |
Cellular Retinoic Acid-binding Protein II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.55 |
Binding ≤ 10μM
|
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Binding ≤ 10μM
|
|
RARB-2-E |
Retinoic Acid Receptor Beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Binding ≤ 10μM
|
|
RARG-2-E |
Retinoic Acid Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
82 |
0.45 |
Binding ≤ 10μM
|
|
RXRB-2-E |
Retinoid X Receptor Beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
306 |
0.41 |
Binding ≤ 10μM
|
|
RXRG-1-E |
Retinoid X Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.41 |
Binding ≤ 10μM |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Functional ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.45 |
Functional ≤ 10μM |
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Functional ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.38 |
Functional ≤ 10μM |
|
RXRB-1-E |
Retinoid X Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Functional ≤ 10μM
|
|
RXRG-2-E |
Retinoid X Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1225 |
0.38 |
Functional ≤ 10μM
|
|
Z50518-2-O |
Human Herpesvirus 4 (cluster #2 Of 5), Other |
Other |
15 |
0.50 |
Functional ≤ 10μM
|
|
Z50594-7-O |
Mus Musculus (cluster #7 Of 9), Other |
Other |
31 |
0.48 |
Functional ≤ 10μM
|
|
Z80135-1-O |
F9 (cluster #1 Of 1), Other |
Other |
200 |
0.43 |
Functional ≤ 10μM |
|
Z80156-5-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #5 Of 12), Other |
Other |
5 |
0.53 |
Functional ≤ 10μM
|
|
Z80166-10-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #10 Of 12), Other |
Other |
4300 |
0.34 |
Functional ≤ 10μM
|
|
Z80224-2-O |
MCF7 (Breast Carcinoma Cells) (cluster #2 Of 14), Other |
Other |
585 |
0.40 |
Functional ≤ 10μM
|
|
Z80377-2-O |
PaCa2 (cluster #2 Of 2), Other |
Other |
9500 |
0.32 |
Functional ≤ 10μM
|
|
Z80390-8-O |
PC-3 (Prostate Carcinoma Cells) (cluster #8 Of 10), Other |
Other |
2000 |
0.36 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
2 |
0.55 |
Functional ≤ 10μM
|
|
Z80712-1-O |
T47D (Breast Carcinoma Cells) (cluster #1 Of 7), Other |
Other |
6 |
0.52 |
Functional ≤ 10μM
|
|
Z81170-2-O |
LNCaP (Prostate Carcinoma) (cluster #2 Of 5), Other |
Other |
500 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.80 |
11.61 |
-54.04 |
0 |
2 |
-1 |
40 |
299.434 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.80 |
11.03 |
-5.3 |
1 |
2 |
0 |
37 |
300.442 |
5 |
↓
|
|