|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
5.51 |
-18.21 |
3 |
5 |
0 |
78 |
390.221 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
-1.04 |
-9.62 |
1 |
2 |
0 |
29 |
295.507 |
2 |
↓
|
|
|
|
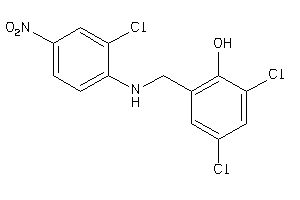
Analogs
-
34659298
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
JAK2-1-E |
Tyrosine-protein Kinase JAK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM |
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM |
|
STAT3-1-E |
Signal Transducer And Activator Of Transcription 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
250 |
0.44 |
Binding ≤ 10μM |
|
Z100499-1-O |
SARS Coronavirus (cluster #1 Of 2), Other |
Other |
100 |
0.47 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
8800 |
0.34 |
Functional ≤ 10μM |
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
6850 |
0.34 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
794 |
0.41 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
700 |
0.41 |
Functional ≤ 10μM |
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
7200 |
0.34 |
Functional ≤ 10μM |
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
700 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
400 |
0.43 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
400 |
0.43 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1400 |
0.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.54 |
6.79 |
-7.83 |
2 |
6 |
0 |
95 |
327.123 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.54 |
7.79 |
-39.71 |
1 |
6 |
-1 |
98 |
326.115 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
-0.66 |
-50.22 |
2 |
4 |
1 |
46 |
328.823 |
4 |
↓
|
|
|
|
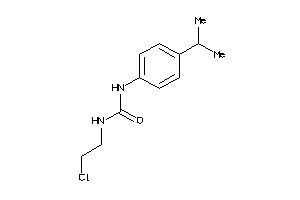
Analogs
-
13558395
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80035-2-O |
B16 (Melanoma Cells) (cluster #2 Of 7), Other |
Other |
8000 |
0.42 |
Functional ≤ 10μM
|
|
Z80088-3-O |
CHO (Ovarian Cells) (cluster #3 Of 4), Other |
Other |
7600 |
0.42 |
Functional ≤ 10μM
|
|
Z80096-2-O |
CHO-TAX 5-6 (cluster #2 Of 2), Other |
Other |
3100 |
0.45 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
9100 |
0.42 |
Functional ≤ 10μM
|
|
Z80186-11-O |
K562 (Erythroleukemia Cells) (cluster #11 Of 11), Other |
Other |
3600 |
0.45 |
Functional ≤ 10μM
|
|
Z80193-10-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #10 Of 12), Other |
Other |
3300 |
0.45 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
4100 |
0.44 |
Functional ≤ 10μM
|
|
Z80362-3-O |
P388 (Lymphoma Cells) (cluster #3 Of 8), Other |
Other |
5200 |
0.44 |
Functional ≤ 10μM
|
|
Z80532-3-O |
T-24 (Bladder Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
6300 |
0.43 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
6200 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.76 |
-1.33 |
-9.94 |
2 |
3 |
0 |
41 |
254.761 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
-2.34 |
-10.99 |
2 |
3 |
0 |
41 |
248.713 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
-2.61 |
-10.36 |
2 |
3 |
0 |
41 |
238.718 |
3 |
↓
|
|
|
|
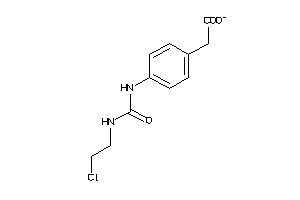
Analogs
-
1704141
-
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
-2.01 |
-10.08 |
2 |
3 |
0 |
41 |
226.707 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
4.75 |
-14.02 |
2 |
3 |
0 |
41 |
226.707 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
-3.22 |
-9.82 |
2 |
3 |
0 |
41 |
277.549 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
3.27 |
-16.33 |
1 |
4 |
0 |
49 |
434.483 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
2.34 |
-17.94 |
1 |
4 |
0 |
49 |
434.483 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
9800 |
0.29 |
Functional ≤ 10μM |
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
2000 |
0.33 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.75 |
11.76 |
-38.41 |
3 |
4 |
1 |
51 |
360.268 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.75 |
11.36 |
-6.83 |
2 |
4 |
0 |
50 |
359.26 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.42 |
Binding ≤ 10μM
|
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
169 |
0.38 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.42 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
169 |
0.38 |
Binding ≤ 10μM
|
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
208 |
0.37 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
208 |
0.37 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
208 |
0.37 |
Binding ≤ 10μM
|
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
55 |
0.41 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
55 |
0.41 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
208 |
0.37 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
55 |
0.41 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
169 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
732 |
0.34 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
65 |
0.40 |
Binding ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
639 |
0.35 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
1354 |
0.33 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
630 |
0.35 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
800 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
8.37 |
-18.97 |
2 |
6 |
0 |
78 |
338.411 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
2.17 |
8.31 |
-20.43 |
2 |
6 |
0 |
78 |
338.411 |
5 |
↓
|
|
|
|
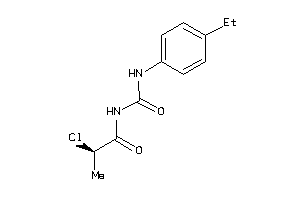
Analogs
-
34963375
-
-
34963377
-
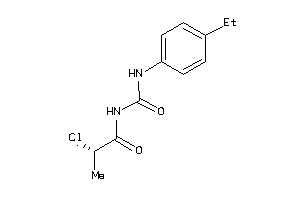
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
6.4 |
-24.77 |
2 |
4 |
0 |
58 |
268.744 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.35 |
Binding ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.35 |
Binding ≤ 10μM
|
|
Z100745-2-O |
MX1 (Breast Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
4 |
0.49 |
Functional ≤ 10μM
|
|
Z80063-1-O |
CCRF S-180 (cluster #1 Of 1), Other |
Other |
30 |
0.44 |
Functional ≤ 10μM
|
|
Z80064-8-O |
CCRF-CEM (T-cell Leukemia) (cluster #8 Of 9), Other |
Other |
753 |
0.36 |
Functional ≤ 10μM
|
|
Z80084-1-O |
CEM-VLB (Vinblastine-resistant T-cell Leukemia Cells) (cluster #1 Of 2), Other |
Other |
3790 |
0.32 |
Functional ≤ 10μM
|
|
Z80085-1-O |
CEM-VM1 (Etoposide-resistant T-cell Leukemia Cells) (cluster #1 Of 2), Other |
Other |
1600 |
0.34 |
Functional ≤ 10μM
|
|
Z80117-2-O |
DC3F/AD-II (cluster #2 Of 2), Other |
Other |
54 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
25 |
0.44 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
880 |
0.35 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
16 |
0.45 |
Functional ≤ 10μM
|
|
Z80555-1-O |
TSGH (cluster #1 Of 1), Other |
Other |
450 |
0.37 |
Functional ≤ 10μM
|
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
47 |
0.43 |
Functional ≤ 10μM
|
|
Z80967-1-O |
Hone-1 Cell Line (cluster #1 Of 2), Other |
Other |
310 |
0.38 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
1400 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
-4.69 |
-10.2 |
4 |
4 |
0 |
71 |
315.376 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.51 |
14.97 |
-138.1 |
6 |
9 |
3 |
96 |
596.8 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.51 |
14.61 |
-93.93 |
5 |
9 |
2 |
95 |
595.792 |
11 |
↓
|
|
|
|
Analogs
-
1539879
-
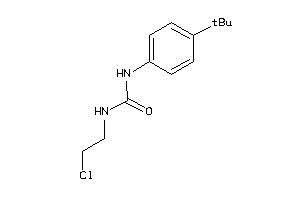
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80035-2-O |
B16 (Melanoma Cells) (cluster #2 Of 7), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z80088-3-O |
CHO (Ovarian Cells) (cluster #3 Of 4), Other |
Other |
3200 |
0.48 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM
|
|
Z80186-11-O |
K562 (Erythroleukemia Cells) (cluster #11 Of 11), Other |
Other |
1900 |
0.50 |
Functional ≤ 10μM
|
|
Z80193-10-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #10 Of 12), Other |
Other |
2000 |
0.50 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
1000 |
0.53 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z80362-3-O |
P388 (Lymphoma Cells) (cluster #3 Of 8), Other |
Other |
2100 |
0.50 |
Functional ≤ 10μM
|
|
Z80532-3-O |
T-24 (Bladder Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
3600 |
0.48 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
2500 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
5.37 |
-9.95 |
2 |
3 |
0 |
41 |
240.734 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
-3.3 |
-9.58 |
2 |
3 |
0 |
41 |
324.549 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
8100 |
0.25 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
9600 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
9.89 |
-16.1 |
4 |
9 |
0 |
118 |
391.435 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
390 |
0.25 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
48 |
0.28 |
Binding ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
48 |
0.28 |
Binding ≤ 10μM
|
|
FGFR3-1-E |
Fibroblast Growth Factor Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
48 |
0.28 |
Binding ≤ 10μM
|
|
FGFR4-1-E |
Fibroblast Growth Factor Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
48 |
0.28 |
Binding ≤ 10μM
|
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.22 |
Binding ≤ 10μM
|
|
MP2K1-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2800 |
0.22 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
360 |
0.25 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
450 |
0.25 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
44 |
0.29 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
450 |
0.25 |
Functional ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
450 |
0.25 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
900 |
0.24 |
Functional ≤ 10μM
|
|
Z80742-1-O |
C6 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
5800 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.21 |
-2.72 |
-50.46 |
4 |
8 |
1 |
96 |
533.528 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
10.69 |
-11.53 |
1 |
5 |
0 |
64 |
389.842 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-3-E |
Serine/threonine-protein Kinase Aurora-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.27 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.18 |
Binding ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
6200 |
0.17 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
640 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.05 |
15.48 |
-21.23 |
3 |
10 |
0 |
120 |
602.07 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
11.47 |
-97.01 |
2 |
3 |
2 |
22 |
291.826 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.25 |
8.71 |
-4.35 |
0 |
3 |
0 |
19 |
289.81 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
1375 |
0.27 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
1400 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
10.06 |
-30.85 |
4 |
7 |
1 |
93 |
419.896 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.19 |
9.69 |
-14.75 |
3 |
7 |
0 |
92 |
418.888 |
6 |
↓
|
|
|
|
Analogs
-
13125756
-
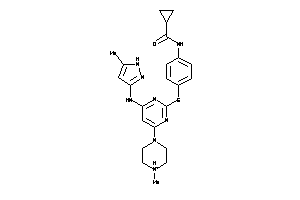
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAK1-1-E |
Adaptor-associated Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.28 |
Binding ≤ 10μM
|
|
AAPK1-1-E |
AMP-activated Protein Kinase, Alpha-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
|
AAPK2-1-E |
AMP-activated Protein Kinase, Alpha-2 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.27 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.34 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.36 |
Binding ≤ 10μM
|
|
ACK1-1-E |
Tyrosine Kinase Non-receptor Protein 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.23 |
Binding ≤ 10μM
|
|
ACVR1-1-E |
Activin Receptor Type-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.24 |
Binding ≤ 10μM
|
|
AKIP-1-E |
Aurora Kinase A-interacting Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Binding ≤ 10μM
|
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.23 |
Binding ≤ 10μM
|
|
ANKK1-1-E |
Ankyrin Repeat And Protein Kinase Domain-containing Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.22 |
Binding ≤ 10μM
|
|
AURKA-1-E |
Serine/threonine-protein Kinase Aurora-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.35 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
740 |
0.26 |
Binding ≤ 10μM
|
|
AURKC-1-E |
Serine/threonine-protein Kinase Aurora-C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.30 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.24 |
Binding ≤ 10μM
|
|
BMP2K-1-E |
BMP-2-inducible Protein Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
65 |
0.30 |
Binding ≤ 10μM
|
|
BMPR2-1-E |
Bone Morphogenetic Protein Receptor Type-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7100 |
0.22 |
Binding ≤ 10μM
|
|
BMX-1-E |
Tyrosine-protein Kinase BMX (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.24 |
Binding ≤ 10μM
|
|
BTK-1-E |
Tyrosine-protein Kinase BTK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.23 |
Binding ≤ 10μM
|
|
CD11A-1-E |
PITSLRE Serine/threonine-protein Kinase CDC2L2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
260 |
0.28 |
Binding ≤ 10μM
|
|
CD11B-1-E |
PITSLRE Serine/threonine-protein Kinase CDC2L1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.28 |
Binding ≤ 10μM
|
|
CDK16-1-E |
Serine/threonine-protein Kinase PCTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.28 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.24 |
Binding ≤ 10μM
|
|
CSK-1-E |
Tyrosine-protein Kinase CSK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.23 |
Binding ≤ 10μM
|
|
CSK21-1-E |
Casein Kinase II Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.27 |
Binding ≤ 10μM
|
|
CSK22-1-E |
Casein Kinase II Alpha (prime) (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.27 |
Binding ≤ 10μM
|
|
DAPK3-1-E |
Death-associated Protein Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.29 |
Binding ≤ 10μM
|
|
DCLK3-1-E |
Serine/threonine-protein Kinase DCLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.23 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.32 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.28 |
Binding ≤ 10μM
|
|
E2AK4-1-E |
Eukaryotic Translation Initiation Factor 2-alpha Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.24 |
Binding ≤ 10μM
|
|
EPHA1-1-E |
Ephrin Type-A Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Binding ≤ 10μM
|
|
EPHA2-1-E |
Ephrin Type-A Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
840 |
0.26 |
Binding ≤ 10μM
|
|
EPHA3-1-E |
Ephrin Type-A Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.25 |
Binding ≤ 10μM
|
|
EPHA4-1-E |
Ephrin Type-A Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.23 |
Binding ≤ 10μM
|
|
EPHA8-1-E |
Ephrin Type-A Receptor 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.27 |
Binding ≤ 10μM
|
|
EPHB1-1-E |
Ephrin Type-B Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.24 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.24 |
Binding ≤ 10μM
|
|
FAK2-1-E |
Protein-tyrosine Kinase 2-beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.24 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
550 |
0.27 |
Binding ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.26 |
Binding ≤ 10μM
|
|
FGFR3-1-E |
Fibroblast Growth Factor Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.26 |
Binding ≤ 10μM
|
|
FGFR4-1-E |
Fibroblast Growth Factor Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.22 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
790 |
0.26 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.35 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.23 |
Binding ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
530 |
0.27 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.23 |
Binding ≤ 10μM
|
|
IGF1R-1-E |
Insulin-like Growth Factor 1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
740 |
0.26 |
Binding ≤ 10μM
|
|
IKKE-1-E |
Inhibitor Of Nuclear Factor Kappa B Kinase Epsilon Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.24 |
Binding ≤ 10μM
|
|
INSR-1-E |
Insulin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
630 |
0.26 |
Binding ≤ 10μM
|
|
INSRR-1-E |
Insulin Receptor-related Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.28 |
Binding ≤ 10μM
|
|
IRAK3-1-E |
Interleukin-1 Receptor-associated Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.22 |
Binding ≤ 10μM
|
|
ITK-1-E |
Tyrosine-protein Kinase ITK/TSK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.27 |
Binding ≤ 10μM
|
|
JAK2-1-E |
Tyrosine-protein Kinase JAK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.29 |
Binding ≤ 10μM
|
|
JAK3-1-E |
Tyrosine-protein Kinase JAK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
630 |
0.26 |
Binding ≤ 10μM
|
|
KCC1D-1-E |
CaM Kinase I Delta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4900 |
0.23 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
430 |
0.27 |
Binding ≤ 10μM
|
|
KS6A2-1-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.24 |
Binding ≤ 10μM
|
|
KS6A3-1-E |
Ribosomal Protein S6 Kinase Alpha 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6400 |
0.22 |
Binding ≤ 10μM
|
|
KSYK-1-E |
Tyrosine-protein Kinase SYK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.24 |
Binding ≤ 10μM
|
|
LATS1-1-E |
Serine/threonine-protein Kinase LATS1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.22 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.30 |
Binding ≤ 10μM
|
|
LIMK1-1-E |
LIM Domain Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.24 |
Binding ≤ 10μM
|
|
LIMK2-1-E |
LIM Domain Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.24 |
Binding ≤ 10μM
|
|
LTK-1-E |
Leukocyte Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7500 |
0.22 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
650 |
0.26 |
Binding ≤ 10μM
|
|
M3K10-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.24 |
Binding ≤ 10μM
|
|
M3K11-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.26 |
Binding ≤ 10μM
|
|
M3K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
M3K9-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 9 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
390 |
0.27 |
Binding ≤ 10μM
|
|
M4K1-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
72 |
0.30 |
Binding ≤ 10μM
|
|
M4K3-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.28 |
Binding ≤ 10μM
|
|
M4K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.24 |
Binding ≤ 10μM
|
|
M4K5-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
83 |
0.30 |
Binding ≤ 10μM
|
|
MARK1-1-E |
Serine/threonine-protein Kinase MARK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.23 |
Binding ≤ 10μM
|
|
MARK2-1-E |
MAP/microtubule Affinity-regulating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.25 |
Binding ≤ 10μM
|
|
MARK3-1-E |
Serine/threonine-protein Kinase C-TAK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
MARK4-1-E |
MAP/microtubule Affinity-regulating Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.23 |
Binding ≤ 10μM
|
|
MELK-1-E |
Maternal Embryonic Leucine Zipper Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.27 |
Binding ≤ 10μM
|
|
MERTK-1-E |
Proto-oncogene Tyrosine-protein Kinase MER (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.24 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
670 |
0.26 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.24 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.24 |
Binding ≤ 10μM
|
|
MP2K1-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.24 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.24 |
Binding ≤ 10μM
|
|
MP2K6-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.24 |
Binding ≤ 10μM
|
|
MUSK-1-E |
Muscle, Skeletal Receptor Tyrosine Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.28 |
Binding ≤ 10μM
|
|
MYLK-1-E |
Myosin Light Chain Kinase, Smooth Muscle (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.33 |
Binding ≤ 10μM
|
|
MYLK2-1-E |
Myosin Light Chain Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.31 |
Binding ≤ 10μM
|
|
MYLK4-1-E |
Myosin Light Chain Kinase Family Member 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.24 |
Binding ≤ 10μM
|
|
NEK5-1-E |
Serine/threonine-protein Kinase Nek5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4200 |
0.23 |
Binding ≤ 10μM
|
|
NTRK1-1-E |
Nerve Growth Factor Receptor Trk-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
38 |
0.31 |
Binding ≤ 10μM
|
|
NTRK2-1-E |
Neurotrophic Tyrosine Kinase Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
240 |
0.28 |
Binding ≤ 10μM
|
|
NTRK3-1-E |
NT-3 Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.28 |
Binding ≤ 10μM
|
|
NUAK1-1-E |
NUAK Family SNF1-like Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.28 |
Binding ≤ 10μM
|
|
NUAK2-1-E |
NUAK Family SNF1-like Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
530 |
0.27 |
Binding ≤ 10μM
|
|
PAK1-1-E |
Serine/threonine-protein Kinase PAK 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.23 |
Binding ≤ 10μM
|
|
PAK2-1-E |
Serine/threonine-protein Kinase PAK 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5700 |
0.22 |
Binding ≤ 10μM
|
|
PAK3-1-E |
Serine/threonine-protein Kinase PAK 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.23 |
Binding ≤ 10μM
|
|
PAK4-1-E |
Serine/threonine-protein Kinase PAK 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.23 |
Binding ≤ 10μM
|
|
PAK7-1-E |
Serine/threonine-protein Kinase PAK7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.23 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.25 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.28 |
Binding ≤ 10μM
|
|
PHKG1-1-E |
Phosphorylase Kinase Gamma Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.24 |
Binding ≤ 10μM
|
|
PI42B-1-E |
Phosphatidylinositol-5-phosphate 4-kinase Type-2 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.29 |
Binding ≤ 10μM
|
|
PI51A-1-E |
Phosphatidylinositol-4-phosphate 5-kinase Type-1 Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
130 |
0.29 |
Binding ≤ 10μM
|
|
PKN1-1-E |
Protein Kinase N1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.25 |
Binding ≤ 10μM
|
|
PLK4-1-E |
Serine/threonine-protein Kinase PLK4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.34 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.32 |
Binding ≤ 10μM
|
|
RIOK1-1-E |
Serine/threonine-protein Kinase RIO1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
360 |
0.27 |
Binding ≤ 10μM
|
|
RIOK3-1-E |
Serine/threonine-protein Kinase RIO3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.26 |
Binding ≤ 10μM
|
|
RIPK1-1-E |
Receptor-interacting Serine/threonine-protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.33 |
Binding ≤ 10μM
|
|
ROS1-1-E |
Proto-oncogene Tyrosine-protein Kinase ROS (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
610 |
0.26 |
Binding ≤ 10μM
|
|
SIK1-1-E |
Serine/threonine-protein Kinase SIK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
SIK2-1-E |
Serine/threonine-protein Kinase SIK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.24 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
82 |
0.30 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
170 |
0.29 |
Binding ≤ 10μM
|
|
SRPK1-1-E |
Serine/threonine-protein Kinase SRPK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
82 |
0.30 |
Binding ≤ 10μM
|
|
SRPK2-1-E |
Serine/threonine-protein Kinase SRPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.26 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.29 |
Binding ≤ 10μM
|
|
STK11-1-E |
Serine/threonine-protein Kinase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.25 |
Binding ≤ 10μM
|
|
STK16-1-E |
Serine/threonine-protein Kinase 16 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
610 |
0.26 |
Binding ≤ 10μM
|
|
STK24-1-E |
Serine/threonine-protein Kinase 24 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.23 |
Binding ≤ 10μM
|
|
STK25-1-E |
Serine/threonine-protein Kinase 25 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.23 |
Binding ≤ 10μM
|
|
STK3-1-E |
Serine/threonine-protein Kinase MST2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
510 |
0.27 |
Binding ≤ 10μM
|
|
STK4-1-E |
Serine/threonine-protein Kinase MST1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.28 |
Binding ≤ 10μM
|
|
TESK1-1-E |
Dual Specificity Testis-specific Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.22 |
Binding ≤ 10μM
|
|
TIE1-1-E |
Tyrosine-protein Kinase Receptor Tie-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.28 |
Binding ≤ 10μM
|
|
TIE2-1-E |
Tyrosine-protein Kinase TIE-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
450 |
0.27 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.24 |
Binding ≤ 10μM
|
|
TNK1-1-E |
Non-receptor Tyrosine-protein Kinase TNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
85 |
0.30 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.24 |
Binding ≤ 10μM
|
|
UFO-1-E |
Tyrosine-protein Kinase Receptor UFO (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
210 |
0.28 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.30 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.24 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.24 |
Binding ≤ 10μM
|
|
WEE1-1-E |
Serine/threonine-protein Kinase WEE1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.23 |
Binding ≤ 10μM
|
|
YES-1-E |
Tyrosine-protein Kinase YES (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.27 |
Binding ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
150 |
0.29 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
48 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
53 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
-6.09 |
-47.27 |
4 |
9 |
1 |
103 |
465.607 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
2100 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
9.37 |
-16.4 |
1 |
3 |
0 |
57 |
306.752 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-6.37 |
24.54 |
-432.11 |
4 |
8 |
6 |
75 |
680.86 |
4 |
↓
|
|
|
|
Analogs
-
38413756
-
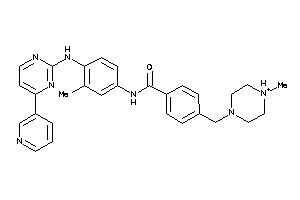
Draw
Identity
99%
90%
80%
70%
Vendors
And 43 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A1Z199-1-E |
BCR/ABL P210 Fusion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
750 |
0.23 |
Binding ≤ 10μM
|
|
ABCG2-2-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4898 |
0.20 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.30 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.31 |
Binding ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
473 |
0.24 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
520 |
0.24 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.21 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
980 |
0.23 |
Binding ≤ 10μM
|
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
7450 |
0.19 |
Binding ≤ 10μM
|
|
CAH14-8-E |
Carbonic Anhydrase XIV (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
468 |
0.24 |
Binding ≤ 10μM
|
|
CAH15-2-E |
Carbonic Anhydrase 15 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
78 |
0.27 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
30 |
0.28 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
528 |
0.24 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
4553 |
0.20 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
392 |
0.24 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
109 |
0.26 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
76 |
0.27 |
Binding ≤ 10μM
|
|
CDK19-1-E |
Cell Division Cycle 2-like Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.20 |
Binding ≤ 10μM
|
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.20 |
Binding ≤ 10μM
|
|
CLK4-1-E |
Dual Specificity Protein Kinase CLK4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.20 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
960 |
0.23 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.28 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.30 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7600 |
0.19 |
Binding ≤ 10μM
|
|
EPHA8-1-E |
Ephrin Type-A Receptor 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.21 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.21 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.21 |
Binding ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5500 |
0.20 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.21 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
97 |
0.27 |
Binding ≤ 10μM
|
|
KSYK-1-E |
Tyrosine-protein Kinase SYK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
62 |
0.27 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.25 |
Binding ≤ 10μM
|
|
MELK-1-E |
Maternal Embryonic Leucine Zipper Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.22 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5200 |
0.20 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.21 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.21 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
72 |
0.27 |
Binding ≤ 10μM
|
|
PLK4-1-E |
Serine/threonine-protein Kinase PLK4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.19 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.22 |
Binding ≤ 10μM
|
|
ST17A-1-E |
Serine/threonine-protein Kinase 17A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5300 |
0.20 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.20 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Functional ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Functional ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Functional ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.27 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
9000 |
0.19 |
Functional ≤ 10μM
|
|
Z80036-1-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #1 Of 3), Other |
Other |
9100 |
0.19 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
60 |
0.27 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
6 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
10000 |
0.19 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
13.15 |
-49.4 |
3 |
8 |
1 |
87 |
494.623 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.89 |
13.03 |
-52.36 |
3 |
8 |
1 |
87 |
494.623 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.89 |
10.78 |
-14.7 |
2 |
8 |
0 |
86 |
493.615 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAH-1-B |
Histone Deacetylase-like Amidohydrolase (cluster #1 Of 2), Bacterial |
Bacteria |
1300 |
0.43 |
Binding ≤ 10μM
|
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.53 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
HDAC1-3-E |
Histone Deacetylase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC2-4-E |
Histone Deacetylase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
921 |
0.44 |
Binding ≤ 10μM |
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC4-2-E |
Histone Deacetylase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
90 |
0.52 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
820 |
0.45 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
65 |
0.53 |
Binding ≤ 10μM
|
|
NCOR2-1-E |
Nuclear Receptor Corepressor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
34 |
0.55 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
Q7K6A1-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.52 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
Q9XYC7-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
59 |
0.53 |
Binding ≤ 10μM
|
|
Q9ZTP8-1-E |
Histone Deacetylase HD1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.55 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
Q94F81-2-E |
Histone Deacetylase HD2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
170 |
0.50 |
Functional ≤ 10μM
|
|
Z103205-2-O |
A431 (cluster #2 Of 4), Other |
Other |
4200 |
0.40 |
Functional ≤ 10μM
|
|
Z103348-2-O |
NCI-H2122 (cluster #2 Of 2), Other |
Other |
7500 |
0.38 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
820 |
0.45 |
Functional ≤ 10μM
|
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
800 |
0.45 |
Functional ≤ 10μM
|
|
Z80054-4-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
8100 |
0.38 |
Functional ≤ 10μM
|
|
Z80064-7-O |
CCRF-CEM (T-cell Leukemia) (cluster #7 Of 9), Other |
Other |
950 |
0.44 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
2120 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-4-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #4 Of 12), Other |
Other |
1200 |
0.44 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
2400 |
0.41 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
2000 |
0.42 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
750 |
0.45 |
Functional ≤ 10μM
|
|
Z80224-6-O |
MCF7 (Breast Carcinoma Cells) (cluster #6 Of 14), Other |
Other |
8500 |
0.37 |
Functional ≤ 10μM
|
|
Z80244-1-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #1 Of 7), Other |
Other |
5000 |
0.39 |
Functional ≤ 10μM
|
|
Z80322-1-O |
NCI-H358 (Lung Carcinama Cells) (cluster #1 Of 2), Other |
Other |
2500 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
6500 |
0.38 |
Functional ≤ 10μM
|
|
Z80418-9-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #9 Of 9), Other |
Other |
760 |
0.45 |
Functional ≤ 10μM
|
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
2100 |
0.42 |
Functional ≤ 10μM
|
|
Z80493-3-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
2500 |
0.41 |
Functional ≤ 10μM
|
|
Z80513-1-O |
SQ20B (cluster #1 Of 1), Other |
Other |
3000 |
0.41 |
Functional ≤ 10μM
|
|
Z80514-1-O |
St-4 (Stomach Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
5200 |
0.39 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
868 |
0.45 |
Functional ≤ 10μM
|
|
Z80838-1-O |
EOL1 (Eosinophilic Cells) (cluster #1 Of 1), Other |
Other |
1000 |
0.44 |
Functional ≤ 10μM
|
|
Z80857-1-O |
Friend Leukemia Cell Line (cluster #1 Of 2), Other |
Other |
990 |
0.44 |
Functional ≤ 10μM
|
|
Z80874-3-O |
CEM (T-cell Leukemia) (cluster #3 Of 7), Other |
Other |
1900 |
0.42 |
Functional ≤ 10μM
|
|
Z80877-1-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #1 Of 2), Other |
Other |
7240 |
0.38 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2060 |
0.42 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
900 |
0.45 |
Functional ≤ 10μM
|
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
9300 |
0.37 |
Functional ≤ 10μM
|
|
Z81024-2-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #2 Of 8), Other |
Other |
3448 |
0.40 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
2600 |
0.41 |
Functional ≤ 10μM
|
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-3-O |
LNCaP (Prostate Carcinoma) (cluster #3 Of 5), Other |
Other |
580 |
0.46 |
Functional ≤ 10μM
|
|
Z81184-5-O |
LOX IMVI (Melanoma Cells) (cluster #5 Of 5), Other |
Other |
1300 |
0.43 |
Functional ≤ 10μM
|
|
Z81245-4-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #4 Of 6), Other |
Other |
1900 |
0.42 |
Functional ≤ 10μM
|
|
Z81247-6-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #6 Of 9), Other |
Other |
460 |
0.47 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
2200 |
0.42 |
Functional ≤ 10μM
|
|
Z81280-3-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #3 Of 3), Other |
Other |
3480 |
0.40 |
Functional ≤ 10μM
|
|
Z81281-4-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #4 Of 5), Other |
Other |
383 |
0.47 |
Functional ≤ 10μM
|
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
832 |
0.45 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
600 |
0.46 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
3900 |
0.40 |
Functional ≤ 10μM
|
|
Z80957-2-O |
HMEC (Microvascular Endothelial Cells) (cluster #2 Of 2), Other |
Other |
1300 |
0.43 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
4.41 |
-18.87 |
3 |
5 |
0 |
78 |
264.325 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.28 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
216 |
0.25 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.28 |
Binding ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
920 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.51 |
15.61 |
-52.14 |
2 |
6 |
1 |
64 |
516.547 |
7 |
↓
|
|