|
|
Analogs
-
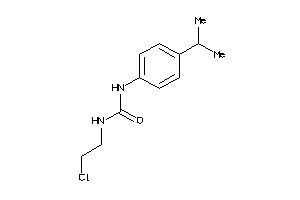
13558395
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80035-2-O |
B16 (Melanoma Cells) (cluster #2 Of 7), Other |
Other |
8000 |
0.42 |
Functional ≤ 10μM
|
|
Z80088-3-O |
CHO (Ovarian Cells) (cluster #3 Of 4), Other |
Other |
7600 |
0.42 |
Functional ≤ 10μM
|
|
Z80096-2-O |
CHO-TAX 5-6 (cluster #2 Of 2), Other |
Other |
3100 |
0.45 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
9100 |
0.42 |
Functional ≤ 10μM
|
|
Z80186-11-O |
K562 (Erythroleukemia Cells) (cluster #11 Of 11), Other |
Other |
3600 |
0.45 |
Functional ≤ 10μM
|
|
Z80193-10-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #10 Of 12), Other |
Other |
3300 |
0.45 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
4100 |
0.44 |
Functional ≤ 10μM
|
|
Z80362-3-O |
P388 (Lymphoma Cells) (cluster #3 Of 8), Other |
Other |
5200 |
0.44 |
Functional ≤ 10μM
|
|
Z80532-3-O |
T-24 (Bladder Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
6300 |
0.43 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
6200 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.76 |
-1.33 |
-9.94 |
2 |
3 |
0 |
41 |
254.761 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
-2.34 |
-10.99 |
2 |
3 |
0 |
41 |
248.713 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
-2.61 |
-10.36 |
2 |
3 |
0 |
41 |
238.718 |
3 |
↓
|
|
|
|
Analogs
-
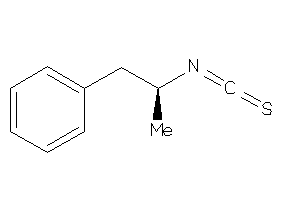
5735314
-
-
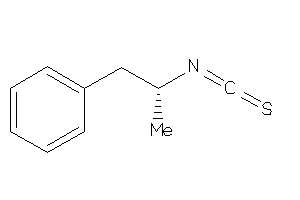
5751594
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.64 |
Binding ≤ 10μM |
|
CP2E1-1-E |
Cytochrome P450 2E1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9980 |
0.64 |
ADME/T ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
9800 |
0.64 |
Functional ≤ 10μM |
|
Z80712-4-O |
T47D (Breast Carcinoma Cells) (cluster #4 Of 7), Other |
Other |
9700 |
0.64 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
5.69 |
-2.35 |
0 |
1 |
0 |
12 |
163.245 |
3 |
↓
|
|
|
|
Analogs
-
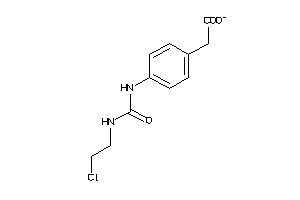
1704141
-
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
-2.01 |
-10.08 |
2 |
3 |
0 |
41 |
226.707 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
4.75 |
-13.93 |
2 |
3 |
0 |
41 |
226.707 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
4.75 |
-14.02 |
2 |
3 |
0 |
41 |
226.707 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
7500 |
0.51 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
5800 |
0.52 |
Functional ≤ 10μM
|
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 9), Other |
Other |
3199 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
1.48 |
-8.47 |
2 |
4 |
0 |
67 |
194.186 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
6.41 |
-13.57 |
2 |
3 |
0 |
41 |
254.761 |
6 |
↓
|
|
|
|
Analogs
-
1539879
-
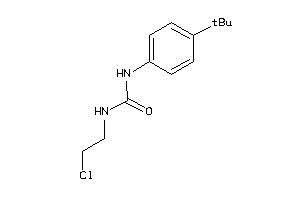
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80035-2-O |
B16 (Melanoma Cells) (cluster #2 Of 7), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z80088-3-O |
CHO (Ovarian Cells) (cluster #3 Of 4), Other |
Other |
3200 |
0.48 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM
|
|
Z80186-11-O |
K562 (Erythroleukemia Cells) (cluster #11 Of 11), Other |
Other |
1900 |
0.50 |
Functional ≤ 10μM
|
|
Z80193-10-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #10 Of 12), Other |
Other |
2000 |
0.50 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
1000 |
0.53 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z80362-3-O |
P388 (Lymphoma Cells) (cluster #3 Of 8), Other |
Other |
2100 |
0.50 |
Functional ≤ 10μM
|
|
Z80532-3-O |
T-24 (Bladder Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
3600 |
0.48 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
2500 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
5.37 |
-9.95 |
2 |
3 |
0 |
41 |
240.734 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
-3.3 |
-9.58 |
2 |
3 |
0 |
41 |
324.549 |
3 |
↓
|
|
|
|
Analogs
-
44136166
-
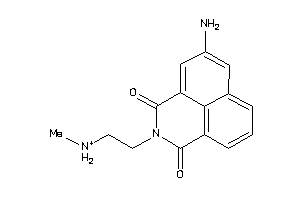
-
3954552
-
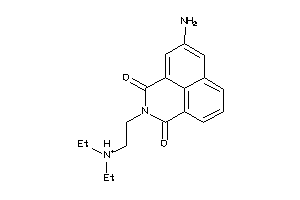
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
7510 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-10-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #10 Of 12), Other |
Other |
1630 |
0.39 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
4670 |
0.36 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
625 |
0.41 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
3500 |
0.36 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
9000 |
0.34 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
2180 |
0.38 |
Functional ≤ 10μM
|
|
Z80362-3-O |
P388 (Lymphoma Cells) (cluster #3 Of 8), Other |
Other |
4560 |
0.36 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
6380 |
0.35 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
4800 |
0.35 |
Functional ≤ 10μM
|
|
Z81017-2-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
2200 |
0.38 |
Functional ≤ 10μM
|
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
6450 |
0.35 |
Functional ≤ 10μM
|
|
Z81024-2-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #2 Of 8), Other |
Other |
7800 |
0.34 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
680 |
0.41 |
Functional ≤ 10μM
|
|
Z81261-1-O |
UACC-375 (cluster #1 Of 1), Other |
Other |
2031 |
0.38 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
8800 |
0.34 |
Functional ≤ 10μM
|
|
Z81341-1-O |
Tumor Cell Lines (cluster #1 Of 1), Other |
Other |
2106 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.11 |
-0.46 |
-34.53 |
3 |
5 |
1 |
69 |
284.339 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
D3ZWV0-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
Q499N1-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.33 |
Binding ≤ 10μM
|
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.33 |
Binding ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.33 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
100 |
0.35 |
Functional ≤ 10μM
|
|
Z80140-1-O |
GBM (cluster #1 Of 1), Other |
Other |
180 |
0.34 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
150 |
0.34 |
Functional ≤ 10μM
|
|
Z80250-1-O |
Me665/2/21 (cluster #1 Of 1), Other |
Other |
250 |
0.33 |
Functional ≤ 10μM
|
|
Z80306-1-O |
NB-4 (Promyelocytic Leukemia Cells) (cluster #1 Of 1), Other |
Other |
82 |
0.35 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
210 |
0.33 |
Functional ≤ 10μM
|
|
Z80449-1-O |
SAOS-2 (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
250 |
0.33 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
260 |
0.33 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
250 |
0.33 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
320 |
0.32 |
Functional ≤ 10μM
|
|
Z81024-5-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #5 Of 8), Other |
Other |
190 |
0.34 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
100 |
0.35 |
Functional ≤ 10μM
|
|
Z81065-3-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
230 |
0.33 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
120 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.53 |
0.19 |
-51.44 |
1 |
3 |
-1 |
60 |
373.472 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
2580 |
0.41 |
Functional ≤ 10μM
|
|
Z103204-1-O |
A427 (cluster #1 Of 4), Other |
Other |
9500 |
0.37 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z50587-3-O |
Homo Sapiens (cluster #3 Of 9), Other |
Other |
5800 |
0.39 |
Functional ≤ 10μM
|
|
Z80035-2-O |
B16 (Melanoma Cells) (cluster #2 Of 7), Other |
Other |
5100 |
0.39 |
Functional ≤ 10μM
|
|
Z80089-1-O |
CHO-AA8 (cluster #1 Of 2), Other |
Other |
36 |
0.55 |
Functional ≤ 10μM
|
|
Z80113-1-O |
Daoy (Medulloblastoma Cells) (cluster #1 Of 2), Other |
Other |
1360 |
0.43 |
Functional ≤ 10μM
|
|
Z80193-10-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #10 Of 12), Other |
Other |
3000 |
0.41 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
9000 |
0.37 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
4500 |
0.39 |
Functional ≤ 10μM
|
|
Z80231-2-O |
MCF7S (Breast Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
1700 |
0.43 |
Functional ≤ 10μM
|
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
790 |
0.45 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
6300 |
0.38 |
Functional ≤ 10μM
|
|
Z80362-3-O |
P388 (Lymphoma Cells) (cluster #3 Of 8), Other |
Other |
7500 |
0.38 |
Functional ≤ 10μM
|
|
Z80578-1-O |
UV4 (cluster #1 Of 1), Other |
Other |
600 |
0.46 |
Functional ≤ 10μM
|
|
Z80768-1-O |
CH1 (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
7900 |
0.38 |
Functional ≤ 10μM
|
|
Z81017-5-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #5 Of 5), Other |
Other |
7130 |
0.38 |
Functional ≤ 10μM
|
|
Z81235-1-O |
M4Beu Cell Line (cluster #1 Of 1), Other |
Other |
5100 |
0.39 |
Functional ≤ 10μM
|
|
Z80362-2-O |
P388 (Lymphoma Cells) (cluster #2 Of 2), Other |
Other |
6750 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
10.01 |
-47.87 |
0 |
3 |
-1 |
43 |
303.209 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.35 |
Binding ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.35 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Binding ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.34 |
Binding ≤ 10μM |
|
Q25270-2-E |
Beta Tubulin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.35 |
Functional ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.38 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.38 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Functional ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4920 |
0.32 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.38 |
Functional ≤ 10μM
|
|
TBB3-3-E |
Tubulin Beta-3 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB4A-3-E |
Tubulin Beta-4 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB4B-3-E |
Tubulin Beta-2 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB8-3-E |
Tubulin Beta-8 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
Z100495-1-O |
BMEC (Brain Microvessel Endothelial Cells) (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z100742-1-O |
MeWo (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM |
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
188 |
0.41 |
Functional ≤ 10μM |
|
Z103208-1-O |
AU565 (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
|
Z103407-2-O |
SW1990 (cluster #2 Of 2), Other |
Other |
250 |
0.40 |
Functional ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
2000 |
0.35 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
18 |
0.47 |
Functional ≤ 10μM
|
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z80013-2-O |
A10 (cluster #2 Of 2), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80035-5-O |
B16 (Melanoma Cells) (cluster #5 Of 7), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z80064-4-O |
CCRF-CEM (T-cell Leukemia) (cluster #4 Of 9), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80071-2-O |
CEM-0 (T-cell Leukemia) (cluster #2 Of 2), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80099-3-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
2756 |
0.34 |
Functional ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
36 |
0.45 |
Functional ≤ 10μM
|
|
Z80121-1-O |
DMS-79 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
124 |
0.42 |
Functional ≤ 10μM
|
|
Z80125-9-O |
DU-145 (Prostate Carcinoma) (cluster #9 Of 9), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z80136-2-O |
FM3A (Breast Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
42 |
0.45 |
Functional ≤ 10μM |
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
94 |
0.43 |
Functional ≤ 10μM |
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM |
|
Z80193-1-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #1 Of 12), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80229-1-O |
MCF7-DOX (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80254-1-O |
MES-SA (Uterine Sarcoma Cells) (cluster #1 Of 2), Other |
Other |
13 |
0.48 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
82 |
0.43 |
Functional ≤ 10μM
|
|
Z80284-3-O |
MOLT-3 (T-lymphoblastic Leukemia Cells) (cluster #3 Of 3), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
16 |
0.47 |
Functional ≤ 10μM
|
|
Z80353-1-O |
OVCAR (cluster #1 Of 1), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
5400 |
0.32 |
Functional ≤ 10μM
|
|
Z80471-3-O |
SH-SY5 (Bone Marrow Neuroblastoma Cells) (cluster #3 Of 3), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM
|
|
Z80485-3-O |
SK-MEL-28 (Melanoma Cells) (cluster #3 Of 6), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM |
|
Z80555-1-O |
TSGH (cluster #1 Of 1), Other |
Other |
45 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80697-3-O |
Bel-7402 (Hepatoma Cells) (cluster #3 Of 3), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80792-1-O |
Colon 26 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z80798-1-O |
CPT30 Cell Line (cluster #1 Of 1), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z80847-3-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #3 Of 3), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80852-2-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z80874-2-O |
CEM (T-cell Leukemia) (cluster #2 Of 7), Other |
Other |
12 |
0.48 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80900-1-O |
HA22T Cell Line (cluster #1 Of 1), Other |
Other |
2708 |
0.34 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM
|
|
Z80967-2-O |
Hone-1 Cell Line (cluster #2 Of 2), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z81001-1-O |
TSGH 9201 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
38 |
0.45 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM
|
|
Z81024-5-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #5 Of 8), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z81065-3-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z81233-2-O |
M21 (Melanoma Cells) (cluster #2 Of 2), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
|
Z81252-7-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #7 Of 11), Other |
Other |
43 |
0.45 |
Functional ≤ 10μM |
|
Z81331-6-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #6 Of 6), Other |
Other |
23 |
0.46 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z100495-1-O |
BMEC (Brain Microvessel Endothelial Cells) (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
ADME/T ≤ 10μM
|
|
Z80013-2-O |
A10 (cluster #2 Of 2), Other |
Other |
23 |
0.46 |
ADME/T ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.52 |
ADME/T ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
ADME/T ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 4), Other |
Other |
2 |
0.53 |
ADME/T ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
ADME/T ≤ 10μM
|
|
Z81057-2-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #2 Of 5), Other |
Other |
5 |
0.51 |
ADME/T ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
3 |
0.52 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
5.32 |
-10.67 |
1 |
5 |
0 |
57 |
316.353 |
6 |
↓
|
|
|
|
Analogs
-
1661
-
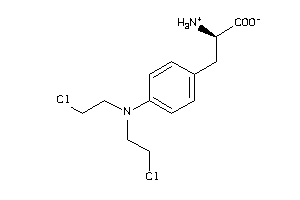
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z103204-1-O |
A427 (cluster #1 Of 4), Other |
Other |
5130 |
0.39 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z50587-3-O |
Homo Sapiens (cluster #3 Of 9), Other |
Other |
3240 |
0.40 |
Functional ≤ 10μM
|
|
Z80008-2-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
310 |
0.48 |
Functional ≤ 10μM
|
|
Z80035-2-O |
B16 (Melanoma Cells) (cluster #2 Of 7), Other |
Other |
2200 |
0.42 |
Functional ≤ 10μM
|
|
Z80064-7-O |
CCRF-CEM (T-cell Leukemia) (cluster #7 Of 9), Other |
Other |
6170 |
0.38 |
Functional ≤ 10μM
|
|
Z80112-1-O |
DAN-G (cluster #1 Of 2), Other |
Other |
2650 |
0.41 |
Functional ≤ 10μM
|
|
Z80156-4-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #4 Of 12), Other |
Other |
2040 |
0.42 |
Functional ≤ 10μM
|
|
Z80193-10-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #10 Of 12), Other |
Other |
981 |
0.44 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
4900 |
0.39 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
5700 |
0.39 |
Functional ≤ 10μM
|
|
Z80231-2-O |
MCF7S (Breast Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
300 |
0.48 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
3240 |
0.40 |
Functional ≤ 10μM
|
|
Z80362-3-O |
P388 (Lymphoma Cells) (cluster #3 Of 8), Other |
Other |
220 |
0.49 |
Functional ≤ 10μM
|
|
Z80473-1-O |
SISO (cluster #1 Of 2), Other |
Other |
1000 |
0.44 |
Functional ≤ 10μM
|
|
Z80604-1-O |
YAPC (cluster #1 Of 2), Other |
Other |
5950 |
0.39 |
Functional ≤ 10μM
|
|
Z80807-1-O |
D283 Cell Line (cluster #1 Of 1), Other |
Other |
6800 |
0.38 |
Functional ≤ 10μM
|
|
Z80874-3-O |
CEM (T-cell Leukemia) (cluster #3 Of 7), Other |
Other |
2500 |
0.41 |
Functional ≤ 10μM
|
|
Z80887-1-O |
H2981 Cell Line (cluster #1 Of 1), Other |
Other |
5000 |
0.39 |
Functional ≤ 10μM
|
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
2200 |
0.42 |
Functional ≤ 10μM
|
|
Z81235-1-O |
M4Beu Cell Line (cluster #1 Of 1), Other |
Other |
1200 |
0.44 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
1900 |
0.42 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
5700 |
0.39 |
Functional ≤ 10μM
|
|
Z81338-1-O |
T-cells (cluster #1 Of 3), Other |
Other |
2200 |
0.42 |
Functional ≤ 10μM
|
|
Z80874-2-O |
CEM (T-cell Leukemia) (cluster #2 Of 4), Other |
Other |
2470 |
0.41 |
ADME/T ≤ 10μM
|
|
Z81325-1-O |
SR (Leukemia Cells) (cluster #1 Of 1), Other |
Other |
1860 |
0.42 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.08 |
6.61 |
-40.9 |
3 |
4 |
0 |
71 |
305.205 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.43 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.43 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.29 |
Functional ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.29 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
1000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.47 |
8.41 |
-93.2 |
5 |
6 |
0 |
109 |
433.599 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.47 |
9.03 |
-87.61 |
5 |
6 |
-1 |
109 |
432.591 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.47 |
8.08 |
-57.51 |
4 |
6 |
-1 |
107 |
432.591 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.24 |
Binding ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2480 |
0.26 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.33 |
Binding ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
280 |
0.31 |
Functional ≤ 10μM
|
|
Z80135-1-O |
F9 (cluster #1 Of 1), Other |
Other |
33 |
0.35 |
Functional ≤ 10μM
|
|
Z80140-1-O |
GBM (cluster #1 Of 1), Other |
Other |
550 |
0.29 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
490 |
0.29 |
Functional ≤ 10μM
|
|
Z80250-1-O |
Me665/2/21 (cluster #1 Of 1), Other |
Other |
420 |
0.30 |
Functional ≤ 10μM
|
|
Z80306-1-O |
NB-4 (Promyelocytic Leukemia Cells) (cluster #1 Of 1), Other |
Other |
400 |
0.30 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
130 |
0.32 |
Functional ≤ 10μM |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
430 |
0.30 |
Functional ≤ 10μM
|
|
Z80449-1-O |
SAOS-2 (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
670 |
0.29 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
440 |
0.30 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
510 |
0.29 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
680 |
0.29 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
430 |
0.30 |
Functional ≤ 10μM
|
|
Z81065-1-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
350 |
0.30 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
320 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.63 |
13.65 |
-51.5 |
1 |
3 |
-1 |
60 |
397.494 |
3 |
↓
|
|
|
|
Analogs
-
34659298
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
JAK2-1-E |
Tyrosine-protein Kinase JAK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM |
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM |
|
STAT3-1-E |
Signal Transducer And Activator Of Transcription 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
250 |
0.44 |
Binding ≤ 10μM |
|
Z100499-1-O |
SARS Coronavirus (cluster #1 Of 2), Other |
Other |
100 |
0.47 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
8800 |
0.34 |
Functional ≤ 10μM |
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
6850 |
0.34 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
794 |
0.41 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
700 |
0.41 |
Functional ≤ 10μM |
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
7200 |
0.34 |
Functional ≤ 10μM |
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
700 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
400 |
0.43 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
400 |
0.43 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1400 |
0.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.54 |
6.79 |
-7.83 |
2 |
6 |
0 |
95 |
327.123 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.54 |
7.79 |
-39.71 |
1 |
6 |
-1 |
98 |
326.115 |
3 |
↓
|
|