|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-1-E |
Heat Shock Protein HSP 90-alpha (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
775 |
0.21 |
Binding ≤ 10μM
|
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
670 |
0.22 |
Binding ≤ 10μM
|
|
HSP90-1-E |
Heat Shock Protein HSP 90 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.20 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.26 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.25 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.25 |
Functional ≤ 10μM
|
|
HS90A-1-E |
Heat Shock Protein HSP 90-alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.25 |
Functional ≤ 10μM
|
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.25 |
Functional ≤ 10μM
|
|
HSP82-1-F |
Heat Shock Protein HSP 90 (HSP82) (cluster #1 Of 1), Fungal |
Fungi |
4800 |
0.19 |
Binding ≤ 10μM
|
|
Z50347-1-O |
Saccharomyces Cerevisiae (cluster #1 Of 1), Other |
Other |
4800 |
0.19 |
Binding ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
200 |
0.23 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
25 |
0.27 |
Functional ≤ 10μM
|
|
Z80186-4-O |
K562 (Erythroleukemia Cells) (cluster #4 Of 11), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z80224-9-O |
MCF7 (Breast Carcinoma Cells) (cluster #9 Of 14), Other |
Other |
9600 |
0.18 |
Functional ≤ 10μM
|
|
Z80262-1-O |
NCI-H1975 (Bronchoalveolar Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
36 |
0.26 |
Functional ≤ 10μM |
|
Z80359-1-O |
P19 (cluster #1 Of 1), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
70 |
0.25 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
30 |
0.26 |
Functional ≤ 10μM |
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
3 |
0.30 |
Functional ≤ 10μM
|
|
Z81186-1-O |
LS174T (Colon Adencocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
450 |
0.22 |
Functional ≤ 10μM
|
|
Z81331-5-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #5 Of 6), Other |
Other |
6 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
3.25 |
-48.59 |
3 |
11 |
-1 |
170 |
559.636 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
1.28 |
0.74 |
-46.33 |
3 |
11 |
-1 |
171 |
559.636 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.88 |
4.88 |
-17.23 |
3 |
11 |
0 |
164 |
560.644 |
5 |
↓
|
|
|
|
Analogs
-
8952080
-
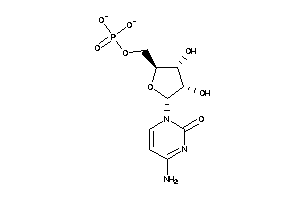
-
9007749
-
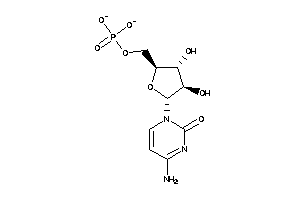
-
16546001
-
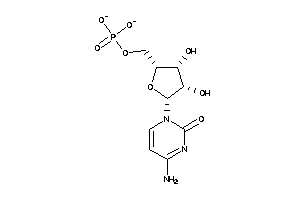
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.60 |
-10.73 |
-143.62 |
4 |
11 |
-2 |
183 |
321.182 |
4 |
↓
|
|
|
|
Analogs
-
3978018
-
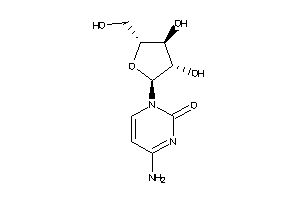
-
6091575
-
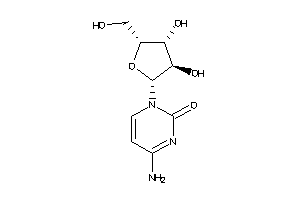
-
6524892
-
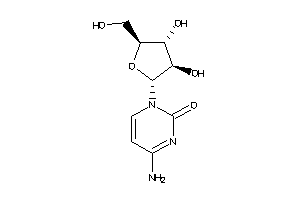
-
16969357
-
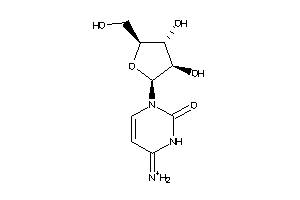
-
38518590
-
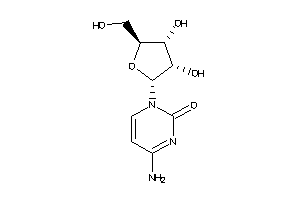
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50515-1-O |
Human Herpesvirus 2 (cluster #1 Of 2), Other |
Other |
2000 |
0.47 |
Functional ≤ 10μM |
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
300 |
0.54 |
Functional ≤ 10μM |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
20 |
0.63 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
90 |
0.58 |
Functional ≤ 10μM
|
|
Z80067-1-O |
CCRF-HSB-2 (T-cell Leukemia Cells) (cluster #1 Of 1), Other |
Other |
150 |
0.56 |
Functional ≤ 10μM
|
|
Z80100-1-O |
COLO 320 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
40 |
0.61 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
8000 |
0.42 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
530 |
0.52 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
66 |
0.59 |
Functional ≤ 10μM |
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
500 |
0.52 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
50 |
0.60 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
100 |
0.58 |
Functional ≤ 10μM
|
|
Z80224-9-O |
MCF7 (Breast Carcinoma Cells) (cluster #9 Of 14), Other |
Other |
620 |
0.51 |
Functional ≤ 10μM
|
|
Z80285-1-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
10 |
0.66 |
Functional ≤ 10μM
|
|
Z80362-6-O |
P388 (Lymphoma Cells) (cluster #6 Of 8), Other |
Other |
140 |
0.56 |
Functional ≤ 10μM
|
|
Z80429-1-O |
RPMI 6410 (cluster #1 Of 1), Other |
Other |
60 |
0.59 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
7680 |
0.42 |
Functional ≤ 10μM
|
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM
|
|
Z80500-1-O |
SNB-7 (cluster #1 Of 1), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM
|
|
Z80678-1-O |
AMML Cell Line (cluster #1 Of 1), Other |
Other |
2300 |
0.46 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
1900 |
0.47 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
2700 |
0.46 |
Functional ≤ 10μM
|
|
Z80777-1-O |
CML/BC Cell Line (cluster #1 Of 1), Other |
Other |
210 |
0.55 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
24 |
0.63 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
5300 |
0.43 |
Functional ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 8), Other |
Other |
200 |
0.55 |
Functional ≤ 10μM
|
|
Z81077-1-O |
KATO III Cell Line (cluster #1 Of 1), Other |
Other |
300 |
0.54 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
700 |
0.51 |
Functional ≤ 10μM
|
|
Z81166-1-O |
LLC Cell Line (cluster #1 Of 1), Other |
Other |
5600 |
0.43 |
Functional ≤ 10μM
|
|
Z81192-1-O |
PC-8 (cluster #1 Of 1), Other |
Other |
1100 |
0.49 |
Functional ≤ 10μM
|
|
Z81193-1-O |
PC-9 (cluster #1 Of 1), Other |
Other |
6600 |
0.43 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
260 |
0.54 |
Functional ≤ 10μM
|
|
Z81281-1-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 5), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.93 |
-7.17 |
-24.49 |
5 |
8 |
0 |
131 |
243.219 |
2 |
↓
|
|
|
|
Analogs
-
22058336
-
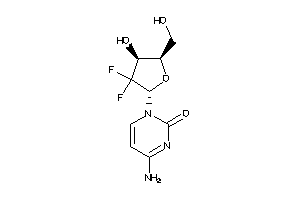
-
6532
-
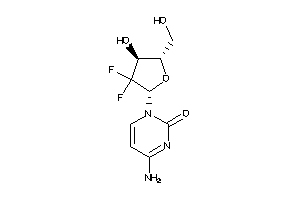
Draw
Identity
99%
90%
80%
70%
Vendors
And 62 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100515-1-O |
HPAC (Pancreatic Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
73 |
0.56 |
Functional ≤ 10μM |
|
Z100733-1-O |
CT26 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
6 |
0.64 |
Functional ≤ 10μM |
|
Z100757-1-O |
SW1573 (Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
8300 |
0.40 |
Functional ≤ 10μM
|
|
Z103393-1-O |
SK-N-AS (cluster #1 Of 1), Other |
Other |
1100 |
0.46 |
Functional ≤ 10μM |
|
Z103407-1-O |
SW1990 (cluster #1 Of 2), Other |
Other |
1200 |
0.46 |
Functional ≤ 10μM
|
|
Z80048-1-O |
BXPC-3 (Pancreatic Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
160 |
0.53 |
Functional ≤ 10μM
|
|
Z80076-1-O |
CEM-SS (T-cell Leukemia) (cluster #1 Of 5), Other |
Other |
30 |
0.59 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
36 |
0.58 |
Functional ≤ 10μM
|
|
Z80132-1-O |
EL4 (Thymoma Cells) (cluster #1 Of 2), Other |
Other |
7 |
0.63 |
Functional ≤ 10μM |
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
330 |
0.50 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
43 |
0.57 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
746 |
0.48 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7 |
0.63 |
Functional ≤ 10μM |
|
Z80224-9-O |
MCF7 (Breast Carcinoma Cells) (cluster #9 Of 14), Other |
Other |
80 |
0.55 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
9 |
0.63 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
40 |
0.58 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
47 |
0.57 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
10 |
0.62 |
Functional ≤ 10μM
|
|
Z80478-1-O |
SK-LU-1 (cluster #1 Of 1), Other |
Other |
55 |
0.56 |
Functional ≤ 10μM
|
|
Z80482-2-O |
SK-MEL-2 (Melanoma Cells) (cluster #2 Of 4), Other |
Other |
7110 |
0.40 |
Functional ≤ 10μM |
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
1700 |
0.45 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
91 |
0.55 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
130 |
0.54 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
90 |
0.55 |
Functional ≤ 10μM
|
|
Z80702-1-O |
BG-1 Cell Line (cluster #1 Of 1), Other |
Other |
680 |
0.48 |
Functional ≤ 10μM
|
|
Z80726-1-O |
TRAMP-C1A (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
9 |
0.63 |
Functional ≤ 10μM
|
|
Z80730-1-O |
C2D Cell Line (cluster #1 Of 1), Other |
Other |
12 |
0.62 |
Functional ≤ 10μM
|
|
Z80731-1-O |
C2G Cell Line (cluster #1 Of 1), Other |
Other |
4 |
0.65 |
Functional ≤ 10μM
|
|
Z80732-1-O |
TRAMP-C2H (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
5 |
0.65 |
Functional ≤ 10μM
|
|
Z80742-1-O |
C6 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
504 |
0.49 |
Functional ≤ 10μM |
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
130 |
0.54 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
10 |
0.62 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
8 |
0.63 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
35 |
0.58 |
Functional ≤ 10μM
|
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
45 |
0.57 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
512 |
0.49 |
Functional ≤ 10μM |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
8 |
0.63 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
245 |
0.51 |
Functional ≤ 10μM |
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
10 |
0.62 |
Functional ≤ 10μM
|
|
Z80335-1-O |
NHDF (cluster #1 Of 2), Other |
Other |
22 |
0.60 |
ADME/T ≤ 10μM
|
|
Z80704-1-O |
BJ (Foreskin Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
9880 |
0.39 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.60 |
-4.98 |
-22.45 |
4 |
7 |
0 |
111 |
263.2 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3880 |
0.29 |
Binding ≤ 10μM |
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5030 |
0.29 |
Binding ≤ 10μM |
|
Z80224-9-O |
MCF7 (Breast Carcinoma Cells) (cluster #9 Of 14), Other |
Other |
3400 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
13.37 |
-13.46 |
0 |
7 |
0 |
90 |
351.362 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3620 |
0.33 |
Binding ≤ 10μM |
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4770 |
0.32 |
Binding ≤ 10μM |
|
Z80224-9-O |
MCF7 (Breast Carcinoma Cells) (cluster #9 Of 14), Other |
Other |
3050 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
11.73 |
-11.84 |
0 |
7 |
0 |
90 |
335.747 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7830 |
0.32 |
Binding ≤ 10μM |
|
Z80224-9-O |
MCF7 (Breast Carcinoma Cells) (cluster #9 Of 14), Other |
Other |
4410 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
11.21 |
-11.65 |
0 |
7 |
0 |
90 |
301.302 |
7 |
↓
|
|
|
|
|