|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
71 |
0.29 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM
|
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
DRD4-3-E |
Dopamine D4 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7300 |
0.21 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3760 |
0.22 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
990 |
0.25 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
372 |
0.26 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
340 |
0.27 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
18 |
0.32 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.25 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4900 |
0.22 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9700 |
0.21 |
ADME/T ≤ 10μM
|
|
DRD2-7-E |
Dopamine D2 Receptor (cluster #7 Of 24), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
15.68 |
-51.49 |
2 |
4 |
1 |
42 |
462.564 |
7 |
↓
|
|
|
|
Analogs
-
34826692
-
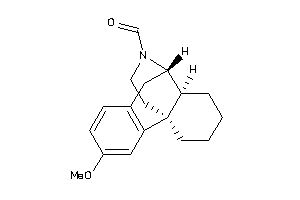
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5070 |
0.37 |
Binding ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3400 |
0.38 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.37 |
ADME/T ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
590 |
0.44 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
2246 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
794 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
8.86 |
-39.89 |
1 |
2 |
1 |
14 |
272.412 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.31 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.31 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.31 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2939 |
0.26 |
Functional ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2939 |
0.26 |
Functional ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2939 |
0.26 |
Functional ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2939 |
0.26 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2939 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
-1 |
-57.15 |
2 |
5 |
1 |
50 |
434.532 |
7 |
↓
|
|
|
|
Analogs
-
13733890
-
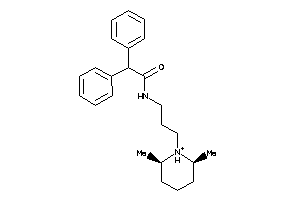
-
13733893
-
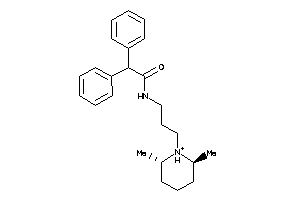
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.38 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.34 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.34 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.38 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.34 |
Binding ≤ 10μM
|
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Functional ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Functional ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
5000 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
12.23 |
-38.7 |
2 |
3 |
1 |
34 |
365.541 |
7 |
↓
|
|
|
|
Analogs
-
13733893
-

-
13733884
-
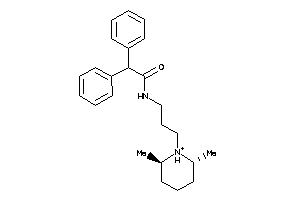
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.38 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.34 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.34 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.38 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.34 |
Binding ≤ 10μM
|
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Functional ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Functional ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
5000 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
12.18 |
-38.1 |
2 |
3 |
1 |
34 |
365.541 |
7 |
↓
|
|
|
|
Analogs
-
13733884
-

-
13733890
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.38 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.34 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.34 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.38 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.34 |
Binding ≤ 10μM
|
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Functional ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Functional ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
5000 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
12.25 |
-38.81 |
2 |
3 |
1 |
34 |
365.541 |
7 |
↓
|
|
|
|
Analogs
-
2753444
-
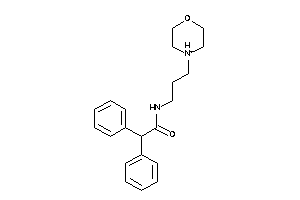
-
2951048
-
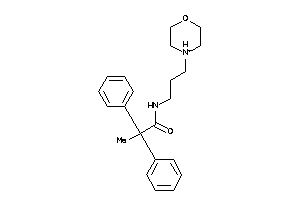
-
19791448
-
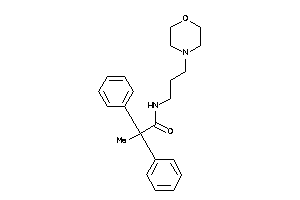
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
6.81 |
-9.73 |
1 |
4 |
0 |
42 |
338.451 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5300 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
0.01 |
-41.46 |
2 |
3 |
1 |
33 |
337.487 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-1-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.33 |
Binding ≤ 10μM
|
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.33 |
Binding ≤ 10μM
|
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.29 |
Binding ≤ 10μM
|
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.25 |
Binding ≤ 10μM
|
|
CAC1I-1-E |
Voltage-gated T-type Calcium Channel Alpha-1I Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
840 |
0.28 |
Binding ≤ 10μM
|
|
CACB1-1-E |
Voltage-dependent L-type Calcium Channel Subunit Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.33 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3630 |
0.25 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9480 |
0.23 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2410 |
0.26 |
Binding ≤ 10μM
|
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.30 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.29 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.29 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.29 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.30 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.29 |
Binding ≤ 10μM
|
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Functional ≤ 10μM
|
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Functional ≤ 10μM
|
|
CAC1I-1-E |
Voltage-gated T-type Calcium Channel Alpha-1I Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Functional ≤ 10μM
|
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.32 |
Functional ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.31 |
Functional ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
420 |
0.30 |
Functional ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.31 |
Functional ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.32 |
Functional ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.31 |
Functional ≤ 10μM
|
|
DRD2-17-E |
Dopamine D2 Receptor (cluster #17 Of 24), Eukaryotic |
Eukaryotes |
228 |
0.31 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.25 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
7300 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.09 |
14.99 |
-46.52 |
1 |
2 |
1 |
8 |
405.512 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.09 |
14.54 |
-45.46 |
1 |
2 |
1 |
8 |
405.512 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.09 |
12.7 |
-5.38 |
0 |
2 |
0 |
6 |
404.504 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
-1.04 |
-40.25 |
1 |
5 |
-1 |
82 |
231.231 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.80 |
1.52 |
-7.19 |
2 |
5 |
0 |
75 |
232.239 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.42 |
-0.99 |
-40.34 |
1 |
5 |
-1 |
82 |
231.231 |
2 |
↓
|
|
|
|
Analogs
-
38572916
-
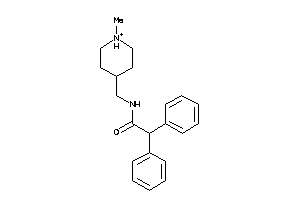
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
12.34 |
-40.9 |
2 |
3 |
1 |
34 |
351.514 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
10.77 |
-40.48 |
2 |
3 |
1 |
34 |
323.46 |
6 |
↓
|
|
|
|
Analogs
-
36731064
-
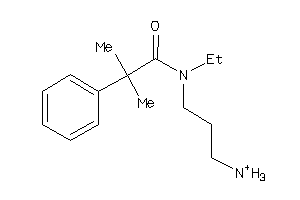
-
1726491
-
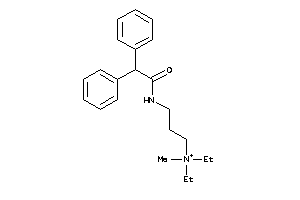
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
0.83 |
-40.82 |
2 |
3 |
1 |
33 |
325.476 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
820 |
0.32 |
Binding ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
820 |
0.32 |
Binding ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
820 |
0.32 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
770 |
0.32 |
Binding ≤ 10μM
|
|
DRD4-3-E |
Dopamine D4 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.44 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.84 |
14.42 |
-46.03 |
1 |
3 |
1 |
30 |
381.927 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NK1R-1-E |
Neurokinin 1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
48 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
9.67 |
-37.5 |
3 |
4 |
1 |
47 |
435.51 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.52 |
8.36 |
-5.3 |
2 |
4 |
0 |
43 |
434.502 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.52 |
9.39 |
-31.02 |
3 |
4 |
1 |
47 |
435.51 |
6 |
↓
|
|
|
|
Analogs
-
26506755
-
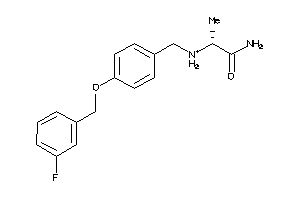
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-3-E |
Monoamine Oxidase B (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
80 |
0.45 |
Binding ≤ 10μM
|
|
SCN1A-2-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A-2-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SCN8A-2-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
20 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
4.1 |
-11.5 |
3 |
4 |
0 |
64 |
302.349 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
SCN9A-1-E |
Sodium Channel Protein Type IX Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Binding ≤ 10μM
|
|
SCN2A-3-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
14 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.42 |
-15.04 |
-31.58 |
10 |
11 |
1 |
192 |
320.278 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNGA1-1-E |
CGMP-gated Cation Channel Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6800 |
0.38 |
Binding ≤ 10μM
|
|
SCN1A-2-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.40 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.40 |
Binding ≤ 10μM
|
|
SCN3A-2-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.40 |
Binding ≤ 10μM
|
|
SCN8A-2-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.40 |
Binding ≤ 10μM
|
|
SCN9A-1-E |
Sodium Channel Protein Type IX Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7510 |
0.38 |
Binding ≤ 10μM
|
|
SCN1A-2-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.47 |
Functional ≤ 10μM |
|
SCN2A-3-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.47 |
Functional ≤ 10μM |
|
SCN3A-2-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.47 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
8.62 |
-37.54 |
2 |
4 |
1 |
43 |
265.377 |
9 |
↓
|
|
|
|
Analogs
-
32120631
-
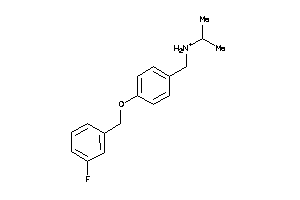
-
53084692
-
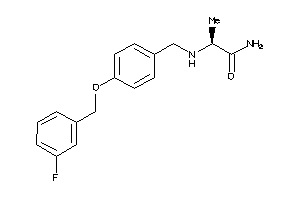
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-3-E |
Monoamine Oxidase B (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
450 |
0.40 |
Binding ≤ 10μM
|
|
SCN1A-2-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8500 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8500 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A-2-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8500 |
0.32 |
Binding ≤ 10μM
|
|
SCN8A-2-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8500 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
280 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
5.36 |
-46.69 |
4 |
4 |
1 |
69 |
303.357 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.91 |
4.13 |
-14.53 |
3 |
4 |
0 |
64 |
302.349 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
16 |
0.30 |
Binding ≤ 10μM
|
|
S29A1-1-E |
Equilibrative Nucleoside Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4954 |
0.21 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2400 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.33 |
15.19 |
-49.33 |
2 |
4 |
1 |
37 |
492.634 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.33 |
12.89 |
-12.84 |
1 |
4 |
0 |
36 |
491.626 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.13 |
8.03 |
-14.25 |
1 |
4 |
0 |
45 |
231.299 |
2 |
↓
|
|
|
|
Analogs
-
40164019
-
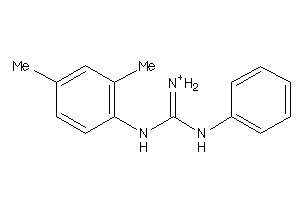
-
40164021
-
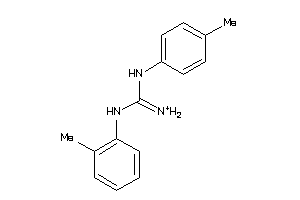
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
744 |
0.48 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2960 |
0.43 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7660 |
0.40 |
Binding ≤ 10μM
|
|
NMD3A-6-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMD3B-6-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE1-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-6-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDZ1-6-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
OPRD-3-E |
Delta Opioid Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
OPRK-2-E |
Kappa Opioid Receptor (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
SGMR1-4-E |
Sigma Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
89 |
0.55 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9210 |
0.39 |
Functional ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
2000 |
0.44 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
70 |
0.56 |
Binding ≤ 10μM
|
|
Z104302-7-O |
Glutamate NMDA Receptor (cluster #7 Of 7), Other |
Other |
9120 |
0.39 |
Binding ≤ 10μM
|
|
Z50512-3-O |
Cavia Porcellus (cluster #3 Of 7), Other |
Other |
6900 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
8.4 |
-25.53 |
4 |
3 |
1 |
50 |
240.33 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1A-1-E |
Voltage-gated P/Q-type Calcium Channel Alpha-1A Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3860 |
0.33 |
Functional ≤ 10μM |
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3860 |
0.33 |
Functional ≤ 10μM |
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3860 |
0.33 |
Functional ≤ 10μM |
|
CAC1E-1-E |
Voltage-gated R-type Calcium Channel Alpha-1E Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3860 |
0.33 |
Functional ≤ 10μM |
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5300 |
0.32 |
Functional ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2550 |
0.34 |
Functional ≤ 10μM |
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5300 |
0.32 |
Functional ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 12), Other |
Other |
7000 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
-3.27 |
-37.36 |
3 |
5 |
1 |
59 |
373.695 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.30 |
-3.11 |
-94.34 |
4 |
5 |
2 |
60 |
374.703 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.30 |
-3.01 |
-90.06 |
4 |
5 |
2 |
60 |
374.703 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
14.8 |
-62.14 |
1 |
4 |
0 |
54 |
473.413 |
9 |
↓
|
|
|
|
Analogs
-
2536946
-
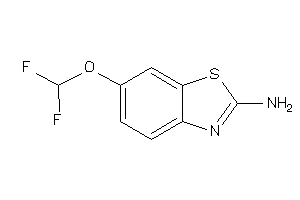
Draw
Identity
99%
90%
80%
70%
Vendors
And 64 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTR1-1-E |
Pteridine Reductase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.50 |
Binding ≤ 10μM |
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.50 |
Functional ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2200 |
0.53 |
Functional ≤ 10μM |
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.50 |
Functional ≤ 10μM
|
|
Z50425-17-O |
Plasmodium Falciparum (cluster #17 Of 22), Other |
Other |
7943 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
2.99 |
-5.52 |
2 |
3 |
0 |
48 |
234.202 |
2 |
↓
|
|