|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.39 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.39 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1197 |
0.30 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.40 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.40 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Functional ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Functional ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Functional ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1197 |
0.30 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1197 |
0.30 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
11 |
0.40 |
Binding ≤ 10μM
|
|
Z80712-1-O |
T47D (Breast Carcinoma Cells) (cluster #1 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80712-1-O |
T47D (Breast Carcinoma Cells) (cluster #1 Of 7), Other |
Other |
1 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
11.72 |
-14.72 |
0 |
4 |
0 |
60 |
386.532 |
3 |
↓
|
|
|
|
Analogs
-
120294
-
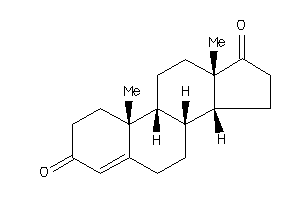
-
2046798
-
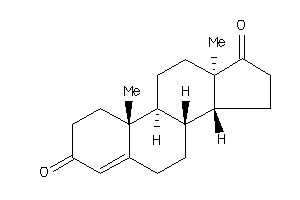
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Binding ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
FABPL-1-E |
Fatty Acid-binding Protein, Liver (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.46 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
31 |
0.46 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.48 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
SGMR1-2-E |
Sigma Opioid Receptor (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
260 |
0.40 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.30 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.30 |
Functional ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2770 |
0.34 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Functional ≤ 10μM
|
|
CP2C9-2-E |
Cytochrome P450 2C9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5500 |
0.32 |
ADME/T ≤ 10μM
|
|
ERG2-1-F |
C-8 Sterol Isomerase (cluster #1 Of 2), Fungal |
Fungi |
4430 |
0.33 |
Binding ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
6310 |
0.32 |
Functional ≤ 10μM
|
|
Z80712-1-O |
T47D (Breast Carcinoma Cells) (cluster #1 Of 7), Other |
Other |
1 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
10.64 |
-9.9 |
0 |
2 |
0 |
34 |
314.469 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
500 |
0.37 |
Functional ≤ 10μM
|
|
Z80017-1-O |
A2780cisR (Cisplatin-resistant Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
60 |
0.42 |
Functional ≤ 10μM
|
|
Z80055-1-O |
CAKI-1 (Kidney Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
40 |
0.43 |
Functional ≤ 10μM
|
|
Z80089-1-O |
CHO-AA8 (cluster #1 Of 2), Other |
Other |
1 |
0.53 |
Functional ≤ 10μM
|
|
Z80114-1-O |
Daudi (Burkitts Lymphoma Cells) (cluster #1 Of 4), Other |
Other |
450 |
0.37 |
Functional ≤ 10μM
|
|
Z80115-1-O |
DC3F (cluster #1 Of 1), Other |
Other |
82 |
0.41 |
Functional ≤ 10μM
|
|
Z80117-1-O |
DC3F/AD-II (cluster #1 Of 2), Other |
Other |
110 |
0.41 |
Functional ≤ 10μM
|
|
Z80125-8-O |
DU-145 (Prostate Carcinoma) (cluster #8 Of 9), Other |
Other |
170 |
0.39 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
50 |
0.43 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
99 |
0.41 |
Functional ≤ 10μM |
|
Z80186-4-O |
K562 (Erythroleukemia Cells) (cluster #4 Of 11), Other |
Other |
470 |
0.37 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
90 |
0.41 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
960 |
0.35 |
Functional ≤ 10μM
|
|
Z80244-3-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #3 Of 7), Other |
Other |
270 |
0.38 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
51 |
0.43 |
Functional ≤ 10μM
|
|
Z80364-1-O |
P388/ADR (Lymphoma Cells) (cluster #1 Of 1), Other |
Other |
535 |
0.37 |
Functional ≤ 10μM
|
|
Z80367-1-O |
P388/S (cluster #1 Of 1), Other |
Other |
48 |
0.43 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
1500 |
0.34 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
200 |
0.39 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
993 |
0.35 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
200 |
0.39 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
7 |
0.48 |
Functional ≤ 10μM
|
|
Z80647-1-O |
833K Cell Line (cluster #1 Of 1), Other |
Other |
328 |
0.38 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
4500 |
0.31 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
400 |
0.37 |
Functional ≤ 10μM
|
|
Z80768-1-O |
CH1 (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
40 |
0.43 |
Functional ≤ 10μM
|
|
Z80776-1-O |
Clone 62 Cell Line (cluster #1 Of 1), Other |
Other |
1500 |
0.34 |
Functional ≤ 10μM
|
|
Z80888-1-O |
H2987 Cell Line (cluster #1 Of 1), Other |
Other |
1600 |
0.34 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
1700 |
0.34 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
10 |
0.47 |
Functional ≤ 10μM
|
|
Z81020-2-O |
HepG2 (Hepatoblastoma Cells) (cluster #2 Of 8), Other |
Other |
5400 |
0.31 |
Functional ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
50 |
0.43 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
600 |
0.36 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
810 |
0.36 |
Functional ≤ 10μM
|
|
Z81264-1-O |
V79 (Lung Fibroblasts) (cluster #1 Of 2), Other |
Other |
800 |
0.36 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
70 |
0.42 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 2), Other |
Other |
4030 |
0.31 |
ADME/T ≤ 10μM
|
|
Z81264-1-O |
V79 (Lung Fibroblasts) (cluster #1 Of 2), Other |
Other |
400 |
0.37 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.23 |
-4.19 |
-10.5 |
4 |
9 |
0 |
145 |
334.332 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.23 |
-4.19 |
-11.87 |
4 |
9 |
0 |
145 |
334.332 |
4 |
↓
|
|
|
|
Analogs
-
4212843
-
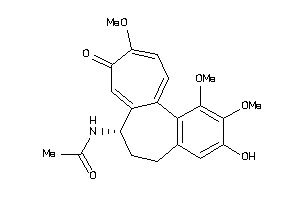
-
607790
-
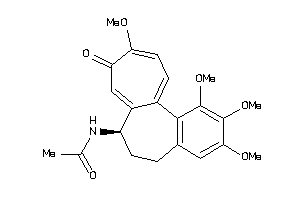
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q862F3-2-E |
Tubulin Alpha Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM |
|
Q862L2-2-E |
Tubulin Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM |
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.27 |
Binding ≤ 10μM
|
|
TBB-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1260 |
0.28 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.29 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.29 |
Binding ≤ 10μM
|
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.30 |
Binding ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.30 |
Binding ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.30 |
Binding ≤ 10μM
|
|
TBG1-1-E |
Tubulin Gamma-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.24 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.24 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.29 |
Functional ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.28 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.29 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.24 |
Functional ≤ 10μM
|
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.26 |
Functional ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
22 |
0.37 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM |
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
53 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
1400 |
0.28 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
18 |
0.37 |
Functional ≤ 10μM |
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
12 |
0.38 |
Functional ≤ 10μM
|
|
Z80024-1-O |
A9 (Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.36 |
Functional ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
31 |
0.36 |
Functional ≤ 10μM
|
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
220 |
0.32 |
Functional ≤ 10μM |
|
Z80096-1-O |
CHO-TAX 5-6 (cluster #1 Of 2), Other |
Other |
140 |
0.33 |
Functional ≤ 10μM |
|
Z80097-1-O |
CHO-VV 3-2 (cluster #1 Of 2), Other |
Other |
620 |
0.30 |
Functional ≤ 10μM |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
190 |
0.32 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
10 |
0.39 |
Functional ≤ 10μM |
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
53 |
0.35 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
1 |
0.43 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
22 |
0.37 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1900 |
0.28 |
Functional ≤ 10μM |
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
15 |
0.38 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
800 |
0.29 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
10 |
0.39 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
33 |
0.36 |
Functional ≤ 10μM |
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM
|
|
Z80427-1-O |
RKO (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM |
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
71 |
0.35 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
30 |
0.36 |
Functional ≤ 10μM |
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
390 |
0.31 |
Functional ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
45 |
0.35 |
Functional ≤ 10μM
|
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
1113 |
0.29 |
Functional ≤ 10μM |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
9 |
0.39 |
Functional ≤ 10μM
|
|
Z80720-1-O |
Burkitts Lymphoma Cells (cluster #1 Of 1), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
52 |
0.35 |
Functional ≤ 10μM |
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
3 |
0.41 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
8 |
0.39 |
Functional ≤ 10μM |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
40 |
0.36 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM |
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
70 |
0.35 |
Functional ≤ 10μM |
|
Z81054-2-O |
SF-268 (Glioblastoma Cells) (cluster #2 Of 3), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
17 |
0.38 |
Functional ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM |
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
93 |
0.34 |
Functional ≤ 10μM |
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
39 |
0.36 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
70 |
0.35 |
Functional ≤ 10μM
|
|
Z81281-1-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 5), Other |
Other |
13 |
0.38 |
Functional ≤ 10μM |
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM |
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM |
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
140 |
0.33 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 3), Other |
Other |
530 |
0.30 |
ADME/T ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
522 |
0.30 |
ADME/T ≤ 10μM
|
|
Z80897-1-O |
H9 (T-lymphoid Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.39 |
ADME/T ≤ 10μM |
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.41 |
ADME/T ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 3), Other |
Other |
210 |
0.32 |
ADME/T ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
2 |
0.42 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
7.99 |
-20.74 |
1 |
7 |
0 |
83 |
399.443 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.45 |
Binding ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
378 |
0.37 |
Binding ≤ 10μM
|
|
KC1A-1-E |
Casein Kinase I Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.30 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.30 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.30 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.30 |
Binding ≤ 10μM
|
|
KC1G2-1-E |
Casein Kinase I Gamma 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.30 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.41 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
45 |
0.43 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
185 |
0.39 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.32 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.32 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.32 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.50 |
Functional ≤ 10μM
|
|
Z100490-1-O |
ASPC1 (Pancreatic Tumour Cell Line) (cluster #1 Of 1), Other |
Other |
592 |
0.36 |
Functional ≤ 10μM
|
|
Z80018-2-O |
A-375 (Malignant Melanoma Cells) (cluster #2 Of 4), Other |
Other |
7 |
0.48 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
17 |
0.45 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
27 |
0.44 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
32 |
0.44 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
10 |
0.47 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
4870 |
0.31 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
9 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
7.83 |
-10.84 |
2 |
5 |
0 |
59 |
325.343 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.95 |
7.96 |
-9.94 |
2 |
5 |
0 |
59 |
325.343 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.95 |
7.89 |
-35.59 |
3 |
5 |
1 |
60 |
326.351 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
10.7 |
-14.87 |
0 |
2 |
0 |
6 |
320.574 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80019-1-O |
A-427 (Lung Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
60 |
0.44 |
Functional ≤ 10μM
|
|
Z80179-1-O |
J82 (Bladder Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
500 |
0.38 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
600 |
0.38 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
90 |
0.43 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
80 |
0.43 |
Functional ≤ 10μM
|
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
60 |
0.44 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
600 |
0.38 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
500 |
0.38 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
400 |
0.39 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
60 |
0.44 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
100 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
-2.17 |
-15.46 |
0 |
4 |
0 |
55 |
362.186 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80019-1-O |
A-427 (Lung Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
100 |
0.41 |
Functional ≤ 10μM
|
|
Z80179-1-O |
J82 (Bladder Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.41 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
900 |
0.35 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
80 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
80 |
0.41 |
Functional ≤ 10μM
|
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
60 |
0.42 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
80 |
0.41 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
70 |
0.42 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
80 |
0.41 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
600 |
0.36 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
50 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
-1.36 |
-15.58 |
0 |
5 |
0 |
64 |
313.316 |
1 |
↓
|
|
|
|
Analogs
-
5735314
-
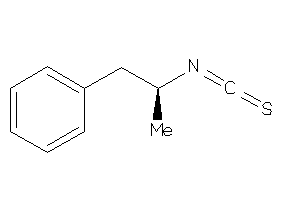
-
5751594
-
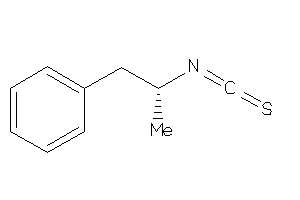
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.64 |
Binding ≤ 10μM |
|
CP2E1-1-E |
Cytochrome P450 2E1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9980 |
0.64 |
ADME/T ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
9800 |
0.64 |
Functional ≤ 10μM |
|
Z80712-4-O |
T47D (Breast Carcinoma Cells) (cluster #4 Of 7), Other |
Other |
9700 |
0.64 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
5.69 |
-2.35 |
0 |
1 |
0 |
12 |
163.245 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.30 |
5.16 |
-16.72 |
2 |
10 |
0 |
127 |
377.43 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
8.26 |
-46.51 |
0 |
8 |
-1 |
105 |
421.458 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.34 |
8.25 |
-16.49 |
1 |
8 |
0 |
103 |
422.466 |
7 |
↓
|
|
|
|
Analogs
-
3917708
-
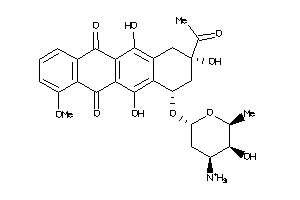
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-1-E |
Serine/threonine-protein Kinase Aurora-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.27 |
Binding ≤ 10μM |
|
MMP2-2-E |
72 KDa Type IV Collagenase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.21 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.22 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 2), Other |
Other |
2600 |
0.20 |
Binding ≤ 10μM
|
|
Z103196-1-O |
22RV1 (cluster #1 Of 2), Other |
Other |
1300 |
0.21 |
Functional ≤ 10μM |
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
130 |
0.25 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
370 |
0.23 |
Functional ≤ 10μM
|
|
Z103219-1-O |
Ca9-22 (cluster #1 Of 1), Other |
Other |
1300 |
0.21 |
Functional ≤ 10μM
|
|
Z50347-3-O |
Saccharomyces Cerevisiae (cluster #3 Of 3), Other |
Other |
10000 |
0.18 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z80008-1-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80017-1-O |
A2780cisR (Cisplatin-resistant Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
6 |
0.30 |
Functional ≤ 10μM
|
|
Z80018-2-O |
A-375 (Malignant Melanoma Cells) (cluster #2 Of 4), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM |
|
Z80021-3-O |
A498 (Renal Carcinoma Cells) (cluster #3 Of 5), Other |
Other |
53 |
0.26 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
700 |
0.22 |
Functional ≤ 10μM |
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
310 |
0.23 |
Functional ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
8 |
0.29 |
Functional ≤ 10μM
|
|
Z80039-1-O |
BGC-823 (Stomach Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
1000 |
0.22 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80068-1-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
6000 |
0.19 |
Functional ≤ 10μM
|
|
Z80092-1-O |
CHO-K1 (Ovarian Cells) (cluster #1 Of 2), Other |
Other |
400 |
0.23 |
Functional ≤ 10μM
|
|
Z80105-2-O |
COR-L23 (Lung Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
319 |
0.23 |
Functional ≤ 10μM
|
|
Z80114-1-O |
Daudi (Burkitts Lymphoma Cells) (cluster #1 Of 4), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
2540 |
0.20 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
6240 |
0.19 |
Functional ≤ 10μM |
|
Z80126-2-O |
DuPro (cluster #2 Of 2), Other |
Other |
1800 |
0.21 |
Functional ≤ 10μM
|
|
Z80138-1-O |
G-361 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
870 |
0.22 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
920 |
0.22 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
940 |
0.22 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
4700 |
0.19 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
9320 |
0.18 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
92 |
0.25 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
950 |
0.22 |
Functional ≤ 10μM
|
|
Z80175-1-O |
IMR-32 (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
430 |
0.23 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
21 |
0.28 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
900 |
0.22 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
200 |
0.24 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
9700 |
0.18 |
Functional ≤ 10μM
|
|
Z80214-2-O |
M14 (Melanoma Cells) (cluster #2 Of 3), Other |
Other |
820 |
0.22 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
80 |
0.25 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
920 |
0.22 |
Functional ≤ 10μM
|
|
Z80227-1-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM
|
|
Z80240-1-O |
MDA-MB-361 (cluster #1 Of 1), Other |
Other |
33 |
0.27 |
Functional ≤ 10μM
|
|
Z80243-1-O |
MDA-MB-453 (Breast Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
120 |
0.25 |
Functional ≤ 10μM
|
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
7 |
0.29 |
Functional ≤ 10μM
|
|
Z80254-1-O |
MES-SA (Uterine Sarcoma Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
704 |
0.22 |
Functional ≤ 10μM
|
|
Z80267-1-O |
MKN-1 (cluster #1 Of 1), Other |
Other |
640 |
0.22 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
3130 |
0.20 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z80295-6-O |
MT4 (Lymphocytes) (cluster #6 Of 8), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80322-2-O |
NCI-H358 (Lung Carcinama Cells) (cluster #2 Of 2), Other |
Other |
620 |
0.22 |
Functional ≤ 10μM
|
|
Z80330-1-O |
NCI-N87 (cluster #1 Of 1), Other |
Other |
420 |
0.23 |
Functional ≤ 10μM
|
|
Z80354-4-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
35 |
0.27 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
17 |
0.28 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
830 |
0.22 |
Functional ≤ 10μM |
|
Z80364-1-O |
P388/ADR (Lymphoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80367-1-O |
P388/S (cluster #1 Of 1), Other |
Other |
13 |
0.28 |
Functional ≤ 10μM
|
|
Z80389-1-O |
PC-14 (cluster #1 Of 1), Other |
Other |
290 |
0.23 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
680 |
0.22 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
912 |
0.22 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
560 |
0.22 |
Functional ≤ 10μM
|
|
Z80441-1-O |
RXF 944 (cluster #1 Of 1), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
600 |
0.22 |
Functional ≤ 10μM |
|
Z80468-1-O |
SF-295 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
35 |
0.27 |
Functional ≤ 10μM
|
|
Z80472-1-O |
SiHa (Cervical Squamous Cell Carcinoma) (cluster #1 Of 2), Other |
Other |
500 |
0.23 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80477-1-O |
SK_HEP1 (Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z80479-1-O |
SK-MEL (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
1730 |
0.21 |
Functional ≤ 10μM
|
|
Z80481-1-O |
SK-MEL-1 (Metastatic Melanoma Cells) (cluster #1 Of 1), Other |
Other |
3092 |
0.20 |
Functional ≤ 10μM |
|
Z80482-3-O |
SK-MEL-2 (Melanoma Cells) (cluster #3 Of 4), Other |
Other |
97 |
0.25 |
Functional ≤ 10μM
|
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
600 |
0.22 |
Functional ≤ 10μM |
|
Z80490-3-O |
SK-MES-1 (cluster #3 Of 4), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80491-2-O |
SK-N-MC (Neuroepithelioma Cells) (cluster #2 Of 4), Other |
Other |
120 |
0.25 |
Functional ≤ 10μM
|
|
Z80492-1-O |
SK-N-SH (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
1100 |
0.21 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
800 |
0.22 |
Functional ≤ 10μM
|
|
Z80495-1-O |
SK-VLB (cluster #1 Of 1), Other |
Other |
6540 |
0.19 |
Functional ≤ 10μM
|
|
Z80503-1-O |
SNU1 (cluster #1 Of 2), Other |
Other |
4000 |
0.19 |
Functional ≤ 10μM
|
|
Z80509-1-O |
SNU-638 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
690 |
0.22 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
640 |
0.22 |
Functional ≤ 10μM
|
|
Z80559-1-O |
U251 (cluster #1 Of 3), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80560-1-O |
U-251 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80574-2-O |
UMUC3 (cluster #2 Of 2), Other |
Other |
9 |
0.29 |
Functional ≤ 10μM
|
|
Z80581-1-O |
VA13 (cluster #1 Of 1), Other |
Other |
400 |
0.23 |
Functional ≤ 10μM
|
|
Z80581-1-O |
VA13 (cluster #1 Of 1), Other |
Other |
699 |
0.22 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
5000 |
0.19 |
Functional ≤ 10μM
|
|
Z80595-1-O |
WIL2-NS (Lymphoblastoid Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.24 |
Functional ≤ 10μM |
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
44 |
0.26 |
Functional ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
9 |
0.29 |
Functional ≤ 10μM
|
|
Z80640-1-O |
786-0 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
3100 |
0.20 |
Functional ≤ 10μM |
|
Z80661-1-O |
A704 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.20 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
540 |
0.22 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
970 |
0.22 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
1000 |
0.22 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
72 |
0.26 |
Functional ≤ 10μM
|
|
Z80726-1-O |
TRAMP-C1A (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
76 |
0.26 |
Functional ≤ 10μM
|
|
Z80730-1-O |
C2D Cell Line (cluster #1 Of 1), Other |
Other |
47 |
0.26 |
Functional ≤ 10μM
|
|
Z80731-1-O |
C2G Cell Line (cluster #1 Of 1), Other |
Other |
29 |
0.27 |
Functional ≤ 10μM
|
|
Z80732-1-O |
TRAMP-C2H (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80735-1-O |
C38 (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80768-1-O |
CH1 (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
6 |
0.30 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
520 |
0.23 |
Functional ≤ 10μM
|
|
Z80801-2-O |
CRL-7065 Cell Line (cluster #2 Of 2), Other |
Other |
500 |
0.23 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
368 |
0.23 |
Functional ≤ 10μM |
|
Z80846-1-O |
F460pv8/eto Cell Line (cluster #1 Of 2), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80887-1-O |
H2981 Cell Line (cluster #1 Of 1), Other |
Other |
700 |
0.22 |
Functional ≤ 10μM
|
|
Z80889-1-O |
NCI-H322M (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
27 |
0.27 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
6810 |
0.19 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
900 |
0.22 |
Functional ≤ 10μM |
|
Z80943-1-O |
HEL 299 (cluster #1 Of 1), Other |
Other |
1700 |
0.21 |
Functional ≤ 10μM |
|
Z80945-1-O |
HEp-2 (Laryngeal Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
59 |
0.26 |
Functional ≤ 10μM
|
|
Z81005-1-O |
HTB-54 Cell Line (cluster #1 Of 1), Other |
Other |
1100 |
0.21 |
Functional ≤ 10μM
|
|
Z81006-1-O |
Human Breast 401NL Tumor Cell Line (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
620 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
954 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
1290 |
0.21 |
Functional ≤ 10μM |
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
660 |
0.22 |
Functional ≤ 10μM
|
|
Z81025-1-O |
Human Lung LXF 529L Tumor Cell Line (cluster #1 Of 1), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z81026-1-O |
Human Lung LXF 629L Tumor Cell Line (cluster #1 Of 1), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM
|
|
Z81028-1-O |
Human Melanoma Cell Line (cluster #1 Of 1), Other |
Other |
280 |
0.24 |
Functional ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z81043-1-O |
N592 (cluster #1 Of 2), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z81101-1-O |
KBM-3 Cell Line (cluster #1 Of 1), Other |
Other |
26 |
0.27 |
Functional ≤ 10μM
|
|
Z81102-1-O |
KBM-3/DOX Cell Line (cluster #1 Of 1), Other |
Other |
490 |
0.23 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
600 |
0.22 |
Functional ≤ 10μM |
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
7010 |
0.19 |
Functional ≤ 10μM
|
|
Z81133-1-O |
L1210 (1565) (cluster #1 Of 1), Other |
Other |
315 |
0.23 |
Functional ≤ 10μM |
|
Z81134-1-O |
L2987 (Lung Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81164-1-O |
LL Cell Line (cluster #1 Of 2), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81167-1-O |
LLTC Cell Line (cluster #1 Of 1), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
78 |
0.26 |
Functional ≤ 10μM
|
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
42 |
0.26 |
Functional ≤ 10μM
|
|
Z81201-1-O |
HOP-62 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
4 |
0.30 |
Functional ≤ 10μM
|
|
Z81211-1-O |
LXFL529 Cell Line (cluster #1 Of 1), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
79 |
0.25 |
Functional ≤ 10μM |
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
960 |
0.22 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
710 |
0.22 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
730 |
0.22 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
840 |
0.22 |
Functional ≤ 10μM
|
|
Z81257-1-O |
MEL-AC Cell Line (cluster #1 Of 1), Other |
Other |
1200 |
0.21 |
Functional ≤ 10μM
|
|
Z81261-1-O |
UACC-375 (cluster #1 Of 1), Other |
Other |
112 |
0.25 |
Functional ≤ 10μM
|
|
Z81277-2-O |
NCI-N417 (cluster #2 Of 2), Other |
Other |
230 |
0.24 |
Functional ≤ 10μM
|
|
Z81280-1-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
8 |
0.29 |
Functional ≤ 10μM
|
|
Z81284-1-O |
NCI-H647 (cluster #1 Of 2), Other |
Other |
1 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
80 |
0.25 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
820 |
0.22 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
2320 |
0.20 |
Functional ≤ 10μM
|
|
Z81341-1-O |
Tumor Cell Lines (cluster #1 Of 1), Other |
Other |
74 |
0.26 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 2), Other |
Other |
500 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
100 |
0.25 |
ADME/T ≤ 10μM
|
|
Z80005-1-O |
3T3 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
3700 |
0.19 |
ADME/T ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
100 |
0.25 |
ADME/T ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
600 |
0.22 |
ADME/T ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 2), Other |
Other |
60 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
480 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80178-2-O |
J774.A1 (Macrophage Cells) (cluster #2 Of 2), Other |
Other |
20 |
0.28 |
ADME/T ≤ 10μM |
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 3), Other |
Other |
60 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80193-3-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #3 Of 4), Other |
Other |
29 |
0.27 |
ADME/T ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
33 |
0.27 |
ADME/T ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
70 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80253-2-O |
MEF (Embryonic Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
3400 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
300 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2000 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80548-2-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
8 |
0.29 |
ADME/T ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 3), Other |
Other |
7400 |
0.18 |
ADME/T ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
900 |
0.22 |
ADME/T ≤ 10μM
|
|
Z80704-1-O |
BJ (Foreskin Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
2700 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80936-4-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #4 Of 4), Other |
Other |
64 |
0.26 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
420 |
0.23 |
ADME/T ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
ADME/T ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
7 |
0.29 |
ADME/T ≤ 10μM
|
|
Z81057-5-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #5 Of 5), Other |
Other |
30 |
0.27 |
ADME/T ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
400 |
0.23 |
ADME/T ≤ 10μM
|
|
Z81331-2-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
178 |
0.24 |
ADME/T ≤ 10μM
|
|
Q7ZJM1-1-V |
Human Immunodeficiency Virus Type 1 Integrase (cluster #1 Of 6), Viral |
Viruses |
900 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
-0.88 |
-58.15 |
8 |
12 |
1 |
208 |
544.533 |
5 |
↓
|
|
|
|
Analogs
-
3830631
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-1-E |
Serine/threonine-protein Kinase Aurora-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.27 |
Binding ≤ 10μM |
|
MMP2-2-E |
72 KDa Type IV Collagenase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.21 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.22 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 2), Other |
Other |
2600 |
0.20 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
130 |
0.25 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
370 |
0.23 |
Functional ≤ 10μM
|
|
Z103219-1-O |
Ca9-22 (cluster #1 Of 1), Other |
Other |
1300 |
0.21 |
Functional ≤ 10μM
|
|
Z50347-3-O |
Saccharomyces Cerevisiae (cluster #3 Of 3), Other |
Other |
10000 |
0.18 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z80008-1-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80017-1-O |
A2780cisR (Cisplatin-resistant Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
6 |
0.30 |
Functional ≤ 10μM
|
|
Z80018-2-O |
A-375 (Malignant Melanoma Cells) (cluster #2 Of 4), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM |
|
Z80021-3-O |
A498 (Renal Carcinoma Cells) (cluster #3 Of 5), Other |
Other |
53 |
0.26 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
700 |
0.22 |
Functional ≤ 10μM |
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
310 |
0.23 |
Functional ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
8 |
0.29 |
Functional ≤ 10μM
|
|
Z80039-1-O |
BGC-823 (Stomach Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
1000 |
0.22 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80068-1-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
6000 |
0.19 |
Functional ≤ 10μM
|
|
Z80092-1-O |
CHO-K1 (Ovarian Cells) (cluster #1 Of 2), Other |
Other |
400 |
0.23 |
Functional ≤ 10μM
|
|
Z80105-2-O |
COR-L23 (Lung Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
319 |
0.23 |
Functional ≤ 10μM
|
|
Z80114-1-O |
Daudi (Burkitts Lymphoma Cells) (cluster #1 Of 4), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
6240 |
0.19 |
Functional ≤ 10μM |
|
Z80126-2-O |
DuPro (cluster #2 Of 2), Other |
Other |
1800 |
0.21 |
Functional ≤ 10μM
|
|
Z80138-1-O |
G-361 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
870 |
0.22 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
920 |
0.22 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
9320 |
0.18 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
92 |
0.25 |
Functional ≤ 10μM
|
|
Z80175-1-O |
IMR-32 (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
21 |
0.28 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
900 |
0.22 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
9700 |
0.18 |
Functional ≤ 10μM
|
|
Z80214-2-O |
M14 (Melanoma Cells) (cluster #2 Of 3), Other |
Other |
820 |
0.22 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
920 |
0.22 |
Functional ≤ 10μM
|
|
Z80227-1-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM
|
|
Z80240-1-O |
MDA-MB-361 (cluster #1 Of 1), Other |
Other |
33 |
0.27 |
Functional ≤ 10μM
|
|
Z80243-1-O |
MDA-MB-453 (Breast Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
120 |
0.25 |
Functional ≤ 10μM
|
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
7 |
0.29 |
Functional ≤ 10μM
|
|
Z80254-1-O |
MES-SA (Uterine Sarcoma Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
704 |
0.22 |
Functional ≤ 10μM
|
|
Z80267-1-O |
MKN-1 (cluster #1 Of 1), Other |
Other |
640 |
0.22 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
3130 |
0.20 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z80295-6-O |
MT4 (Lymphocytes) (cluster #6 Of 8), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80322-2-O |
NCI-H358 (Lung Carcinama Cells) (cluster #2 Of 2), Other |
Other |
620 |
0.22 |
Functional ≤ 10μM
|
|
Z80330-1-O |
NCI-N87 (cluster #1 Of 1), Other |
Other |
420 |
0.23 |
Functional ≤ 10μM
|
|
Z80354-4-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
35 |
0.27 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
830 |
0.22 |
Functional ≤ 10μM |
|
Z80364-1-O |
P388/ADR (Lymphoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80367-1-O |
P388/S (cluster #1 Of 1), Other |
Other |
13 |
0.28 |
Functional ≤ 10μM
|
|
Z80389-1-O |
PC-14 (cluster #1 Of 1), Other |
Other |
290 |
0.23 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
912 |
0.22 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
560 |
0.22 |
Functional ≤ 10μM
|
|
Z80441-1-O |
RXF 944 (cluster #1 Of 1), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
600 |
0.22 |
Functional ≤ 10μM |
|
Z80468-1-O |
SF-295 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
35 |
0.27 |
Functional ≤ 10μM
|
|
Z80472-1-O |
SiHa (Cervical Squamous Cell Carcinoma) (cluster #1 Of 2), Other |
Other |
500 |
0.23 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80477-1-O |
SK_HEP1 (Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z80479-1-O |
SK-MEL (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
1730 |
0.21 |
Functional ≤ 10μM
|
|
Z80481-1-O |
SK-MEL-1 (Metastatic Melanoma Cells) (cluster #1 Of 1), Other |
Other |
3092 |
0.20 |
Functional ≤ 10μM |
|
Z80482-3-O |
SK-MEL-2 (Melanoma Cells) (cluster #3 Of 4), Other |
Other |
97 |
0.25 |
Functional ≤ 10μM
|
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
600 |
0.22 |
Functional ≤ 10μM |
|
Z80490-3-O |
SK-MES-1 (cluster #3 Of 4), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80491-2-O |
SK-N-MC (Neuroepithelioma Cells) (cluster #2 Of 4), Other |
Other |
120 |
0.25 |
Functional ≤ 10μM
|
|
Z80492-1-O |
SK-N-SH (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
1100 |
0.21 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
800 |
0.22 |
Functional ≤ 10μM
|
|
Z80495-1-O |
SK-VLB (cluster #1 Of 1), Other |
Other |
6540 |
0.19 |
Functional ≤ 10μM
|
|
Z80503-1-O |
SNU1 (cluster #1 Of 2), Other |
Other |
4000 |
0.19 |
Functional ≤ 10μM
|
|
Z80509-1-O |
SNU-638 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
690 |
0.22 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
640 |
0.22 |
Functional ≤ 10μM
|
|
Z80559-1-O |
U251 (cluster #1 Of 3), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80560-1-O |
U-251 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80574-2-O |
UMUC3 (cluster #2 Of 2), Other |
Other |
9 |
0.29 |
Functional ≤ 10μM
|
|
Z80581-1-O |
VA13 (cluster #1 Of 1), Other |
Other |
699 |
0.22 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
5000 |
0.19 |
Functional ≤ 10μM
|
|
Z80595-1-O |
WIL2-NS (Lymphoblastoid Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.24 |
Functional ≤ 10μM |
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
44 |
0.26 |
Functional ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
9 |
0.29 |
Functional ≤ 10μM
|
|
Z80661-1-O |
A704 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.20 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
970 |
0.22 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
1000 |
0.22 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
72 |
0.26 |
Functional ≤ 10μM
|
|
Z80726-1-O |
TRAMP-C1A (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
76 |
0.26 |
Functional ≤ 10μM
|
|
Z80730-1-O |
C2D Cell Line (cluster #1 Of 1), Other |
Other |
47 |
0.26 |
Functional ≤ 10μM
|
|
Z80731-1-O |
C2G Cell Line (cluster #1 Of 1), Other |
Other |
29 |
0.27 |
Functional ≤ 10μM
|
|
Z80732-1-O |
TRAMP-C2H (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80735-1-O |
C38 (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80768-1-O |
CH1 (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
6 |
0.30 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
520 |
0.23 |
Functional ≤ 10μM
|
|
Z80801-2-O |
CRL-7065 Cell Line (cluster #2 Of 2), Other |
Other |
500 |
0.23 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
368 |
0.23 |
Functional ≤ 10μM |
|
Z80846-1-O |
F460pv8/eto Cell Line (cluster #1 Of 2), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80887-1-O |
H2981 Cell Line (cluster #1 Of 1), Other |
Other |
700 |
0.22 |
Functional ≤ 10μM
|
|
Z80889-1-O |
NCI-H322M (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
27 |
0.27 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
900 |
0.22 |
Functional ≤ 10μM |
|
Z80943-1-O |
HEL 299 (cluster #1 Of 1), Other |
Other |
1700 |
0.21 |
Functional ≤ 10μM |
|
Z80945-1-O |
HEp-2 (Laryngeal Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
59 |
0.26 |
Functional ≤ 10μM
|
|
Z81005-1-O |
HTB-54 Cell Line (cluster #1 Of 1), Other |
Other |
1100 |
0.21 |
Functional ≤ 10μM
|
|
Z81006-1-O |
Human Breast 401NL Tumor Cell Line (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
620 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
954 |
0.22 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
660 |
0.22 |
Functional ≤ 10μM
|
|
Z81025-1-O |
Human Lung LXF 529L Tumor Cell Line (cluster #1 Of 1), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z81026-1-O |
Human Lung LXF 629L Tumor Cell Line (cluster #1 Of 1), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM
|
|
Z81028-1-O |
Human Melanoma Cell Line (cluster #1 Of 1), Other |
Other |
280 |
0.24 |
Functional ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z81043-1-O |
N592 (cluster #1 Of 2), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z81101-1-O |
KBM-3 Cell Line (cluster #1 Of 1), Other |
Other |
26 |
0.27 |
Functional ≤ 10μM
|
|
Z81102-1-O |
KBM-3/DOX Cell Line (cluster #1 Of 1), Other |
Other |
490 |
0.23 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
7010 |
0.19 |
Functional ≤ 10μM
|
|
Z81133-1-O |
L1210 (1565) (cluster #1 Of 1), Other |
Other |
315 |
0.23 |
Functional ≤ 10μM |
|
Z81134-1-O |
L2987 (Lung Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81164-1-O |
LL Cell Line (cluster #1 Of 2), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81167-1-O |
LLTC Cell Line (cluster #1 Of 1), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
78 |
0.26 |
Functional ≤ 10μM
|
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
42 |
0.26 |
Functional ≤ 10μM
|
|
Z81201-1-O |
HOP-62 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
4 |
0.30 |
Functional ≤ 10μM
|
|
Z81211-1-O |
LXFL529 Cell Line (cluster #1 Of 1), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
79 |
0.25 |
Functional ≤ 10μM |
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
960 |
0.22 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
730 |
0.22 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM
|
|
Z81257-1-O |
MEL-AC Cell Line (cluster #1 Of 1), Other |
Other |
1200 |
0.21 |
Functional ≤ 10μM
|
|
Z81261-1-O |
UACC-375 (cluster #1 Of 1), Other |
Other |
112 |
0.25 |
Functional ≤ 10μM
|
|
Z81277-2-O |
NCI-N417 (cluster #2 Of 2), Other |
Other |
230 |
0.24 |
Functional ≤ 10μM
|
|
Z81280-1-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
8 |
0.29 |
Functional ≤ 10μM
|
|
Z81284-1-O |
NCI-H647 (cluster #1 Of 2), Other |
Other |
1 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
80 |
0.25 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
820 |
0.22 |
Functional ≤ 10μM
|
|
Z81341-1-O |
Tumor Cell Lines (cluster #1 Of 1), Other |
Other |
74 |
0.26 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 2), Other |
Other |
500 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
100 |
0.25 |
ADME/T ≤ 10μM
|
|
Z80005-1-O |
3T3 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
3700 |
0.19 |
ADME/T ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
100 |
0.25 |
ADME/T ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
600 |
0.22 |
ADME/T ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 2), Other |
Other |
60 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
480 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80178-2-O |
J774.A1 (Macrophage Cells) (cluster #2 Of 2), Other |
Other |
20 |
0.28 |
ADME/T ≤ 10μM |
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 3), Other |
Other |
60 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80193-3-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #3 Of 4), Other |
Other |
29 |
0.27 |
ADME/T ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
33 |
0.27 |
ADME/T ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
70 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80253-2-O |
MEF (Embryonic Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
3400 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
300 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2000 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80548-2-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
8 |
0.29 |
ADME/T ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 3), Other |
Other |
7400 |
0.18 |
ADME/T ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
900 |
0.22 |
ADME/T ≤ 10μM
|
|
Z80704-1-O |
BJ (Foreskin Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
2700 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80936-4-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #4 Of 4), Other |
Other |
64 |
0.26 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
420 |
0.23 |
ADME/T ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
ADME/T ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
7 |
0.29 |
ADME/T ≤ 10μM
|
|
Z81057-5-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #5 Of 5), Other |
Other |
30 |
0.27 |
ADME/T ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
400 |
0.23 |
ADME/T ≤ 10μM
|
|
Z81331-2-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
178 |
0.24 |
ADME/T ≤ 10μM
|
|
Q7ZJM1-1-V |
Human Immunodeficiency Virus Type 1 Integrase (cluster #1 Of 6), Viral |
Viruses |
900 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
-0.9 |
-63.19 |
8 |
12 |
1 |
208 |
544.533 |
5 |
↓
|
|
|
|
Analogs
-
31911781
-
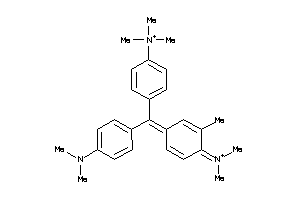
-
3953819
-
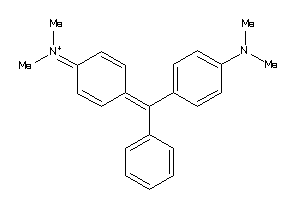
Draw
Identity
99%
90%
80%
70%
Vendors
And 48 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
Z104295-2-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #2 Of 2), Other |
Other |
590 |
0.31 |
Binding ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
2100 |
0.28 |
Functional ≤ 10μM
|
|
Z80021-4-O |
A498 (Renal Carcinoma Cells) (cluster #4 Of 5), Other |
Other |
57 |
0.36 |
Functional ≤ 10μM
|
|
Z80099-3-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
13 |
0.39 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
15 |
0.39 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
14 |
0.39 |
Functional ≤ 10μM
|
|
Z80712-4-O |
T47D (Breast Carcinoma Cells) (cluster #4 Of 7), Other |
Other |
16 |
0.39 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
34 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
13.34 |
-19.69 |
0 |
3 |
1 |
9 |
372.536 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 75 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-3-E |
Thymidylate Synthase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.80 |
Binding ≤ 10μM
|
|
Z103205-4-O |
A431 (cluster #4 Of 4), Other |
Other |
18 |
1.20 |
Functional ≤ 10μM
|
|
Z103419-1-O |
UACC-812 (cluster #1 Of 2), Other |
Other |
3100 |
0.86 |
Functional ≤ 10μM
|
|
Z50425-14-O |
Plasmodium Falciparum (cluster #14 Of 22), Other |
Other |
7943 |
0.79 |
Functional ≤ 10μM
|
|
Z80035-6-O |
B16 (Melanoma Cells) (cluster #6 Of 7), Other |
Other |
8730 |
0.79 |
Functional ≤ 10μM
|
|
Z80054-2-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #2 Of 4), Other |
Other |
1500 |
0.91 |
Functional ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
5000 |
0.82 |
Functional ≤ 10μM
|
|
Z80088-4-O |
CHO (Ovarian Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.82 |
Functional ≤ 10μM
|
|
Z80101-1-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
1100 |
0.93 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
6000 |
0.81 |
Functional ≤ 10μM |
|
Z80125-6-O |
DU-145 (Prostate Carcinoma) (cluster #6 Of 9), Other |
Other |
4800 |
0.83 |
Functional ≤ 10μM
|
|
Z80136-3-O |
FM3A (Breast Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
180 |
1.05 |
Functional ≤ 10μM
|
|
Z80156-6-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #6 Of 12), Other |
Other |
9500 |
0.78 |
Functional ≤ 10μM
|
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
9450 |
0.78 |
Functional ≤ 10μM
|
|
Z80166-8-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #8 Of 12), Other |
Other |
7300 |
0.80 |
Functional ≤ 10μM
|
|
Z80186-7-O |
K562 (Erythroleukemia Cells) (cluster #7 Of 11), Other |
Other |
2400 |
0.87 |
Functional ≤ 10μM
|
|
Z80192-2-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #2 Of 4), Other |
Other |
8320 |
0.79 |
Functional ≤ 10μM
|
|
Z80193-8-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #8 Of 12), Other |
Other |
970 |
0.94 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
9600 |
0.78 |
Functional ≤ 10μM
|
|
Z80268-1-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2500 |
0.87 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3000 |
0.86 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
4400 |
0.83 |
Functional ≤ 10μM |
|
Z80362-5-O |
P388 (Lymphoma Cells) (cluster #5 Of 8), Other |
Other |
609 |
0.97 |
Functional ≤ 10μM |
|
Z80390-6-O |
PC-3 (Prostate Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
16 |
1.21 |
Functional ≤ 10μM
|
|
Z80485-4-O |
SK-MEL-28 (Melanoma Cells) (cluster #4 Of 6), Other |
Other |
4500 |
0.83 |
Functional ≤ 10μM
|
|
Z80490-4-O |
SK-MES-1 (cluster #4 Of 4), Other |
Other |
310 |
1.01 |
Functional ≤ 10μM
|
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
5700 |
0.82 |
Functional ≤ 10μM |
|
Z80526-3-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #3 Of 6), Other |
Other |
8100 |
0.79 |
Functional ≤ 10μM
|
|
Z80532-5-O |
T-24 (Bladder Carcinoma Cells) (cluster #5 Of 6), Other |
Other |
4100 |
0.84 |
Functional ≤ 10μM
|
|
Z80583-6-O |
Vero (Kidney Cells) (cluster #6 Of 7), Other |
Other |
6810 |
0.80 |
Functional ≤ 10μM
|
|
Z80641-1-O |
791T Cell Line (cluster #1 Of 1), Other |
Other |
5500 |
0.82 |
Functional ≤ 10μM
|
|
Z80653-2-O |
9L (Glioma Cells) (cluster #2 Of 2), Other |
Other |
1000 |
0.93 |
Functional ≤ 10μM
|
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
9200 |
0.78 |
Functional ≤ 10μM
|
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
3900 |
0.84 |
Functional ≤ 10μM |
|
Z80712-5-O |
T47D (Breast Carcinoma Cells) (cluster #5 Of 7), Other |
Other |
600 |
0.97 |
Functional ≤ 10μM
|
|
Z80724-1-O |
C170 Cell Line (cluster #1 Of 1), Other |
Other |
2000 |
0.89 |
Functional ≤ 10μM
|
|
Z80742-2-O |
C6 (Glioma Cells) (cluster #2 Of 3), Other |
Other |
6100 |
0.81 |
Functional ≤ 10μM
|
|
Z80874-5-O |
CEM (T-cell Leukemia) (cluster #5 Of 7), Other |
Other |
9230 |
0.78 |
Functional ≤ 10μM
|
|
Z80928-7-O |
HCT-116 (Colon Carcinoma Cells) (cluster #7 Of 9), Other |
Other |
8510 |
0.79 |
Functional ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 8), Other |
Other |
9200 |
0.78 |
Functional ≤ 10μM
|
|
Z81024-6-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #6 Of 8), Other |
Other |
8500 |
0.79 |
Functional ≤ 10μM |
|
Z81034-5-O |
A2780 (Ovarian Carcinoma Cells) (cluster #5 Of 10), Other |
Other |
7210 |
0.80 |
Functional ≤ 10μM
|
|
Z81041-1-O |
Prostatic Carcinoma Cells (cluster #1 Of 2), Other |
Other |
6400 |
0.81 |
Functional ≤ 10μM
|
|
Z81043-2-O |
N592 (cluster #2 Of 2), Other |
Other |
5400 |
0.82 |
Functional ≤ 10μM
|
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3600 |
0.85 |
Functional ≤ 10μM
|
|
Z81115-6-O |
KB (Squamous Cell Carcinoma) (cluster #6 Of 6), Other |
Other |
4460 |
0.83 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
6200 |
0.81 |
Functional ≤ 10μM |
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
4900 |
0.83 |
Functional ≤ 10μM
|
|
Z81247-7-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #7 Of 9), Other |
Other |
9700 |
0.78 |
Functional ≤ 10μM
|
|
Z81252-9-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #9 Of 11), Other |
Other |
9600 |
0.78 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
2700 |
0.87 |
Functional ≤ 10μM |
|
Z81331-3-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #3 Of 6), Other |
Other |
9000 |
0.78 |
Functional ≤ 10μM
|
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
9710 |
0.78 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 4), Other |
Other |
160 |
1.06 |
ADME/T ≤ 10μM
|
|
Z80192-2-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
7940 |
0.79 |
ADME/T ≤ 10μM
|
|
Z80193-4-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #4 Of 4), Other |
Other |
560 |
0.97 |
ADME/T ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
200 |
1.04 |
ADME/T ≤ 10μM
|
|
Z80901-2-O |
HaCaT (Keratinocytes) (cluster #2 Of 2), Other |
Other |
400 |
1.00 |
ADME/T ≤ 10μM
|
|
Z80928-2-O |
HCT-116 (Colon Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
2040 |
0.89 |
ADME/T ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 3), Other |
Other |
570 |
0.97 |
ADME/T ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
6610 |
0.81 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.59 |
-1.23 |
-13.33 |
2 |
4 |
0 |
66 |
130.078 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.13 |
-4 |
-48.58 |
1 |
4 |
-1 |
69 |
129.07 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.13 |
-3.14 |
-32.57 |
1 |
4 |
-1 |
69 |
129.07 |
0 |
↓
|
|
|
|
Analogs
-
576015
-
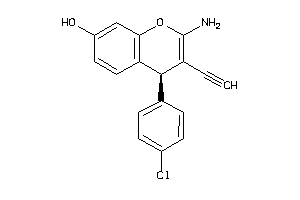
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
80 |
0.45 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
160 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
0.49 |
-8.11 |
2 |
4 |
0 |
68 |
312.756 |
2 |
↓
|
|
|
|
Analogs
-
576015
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
80 |
0.45 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
160 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
0.49 |
-7.51 |
2 |
4 |
0 |
68 |
312.756 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
34004523
-
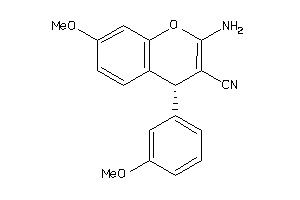
-
34004524
-
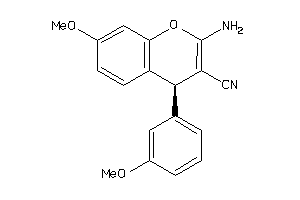
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
15 |
0.44 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
32 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
4.23 |
-11.47 |
2 |
6 |
0 |
87 |
338.363 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.82 |
4.57 |
-49.31 |
3 |
6 |
1 |
88 |
339.371 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
130 |
0.39 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
210 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
-1.56 |
-10.89 |
3 |
6 |
0 |
97 |
403.232 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
130 |
0.39 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
210 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
-1.55 |
-10.87 |
3 |
6 |
0 |
97 |
403.232 |
3 |
↓
|
|
|
|
Analogs
-
1532734
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-3-E |
Carbonic Anhydrase I (cluster #3 Of 12), Eukaryotic |
Eukaryotes |
1400 |
0.82 |
Binding ≤ 10μM
|
|
CAH14-2-E |
Carbonic Anhydrase XIV (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.84 |
Binding ≤ 10μM
|
|
CAH15-3-E |
Carbonic Anhydrase 15 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.70 |
Binding ≤ 10μM
|
|
CAH2-4-E |
Carbonic Anhydrase II (cluster #4 Of 15), Eukaryotic |
Eukaryotes |
1110 |
0.83 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
112 |
0.97 |
Binding ≤ 10μM
|
|
CAH5A-4-E |
Carbonic Anhydrase VA (cluster #4 Of 10), Eukaryotic |
Eukaryotes |
1220 |
0.83 |
Binding ≤ 10μM
|
|
CAH5B-5-E |
Carbonic Anhydrase VB (cluster #5 Of 9), Eukaryotic |
Eukaryotes |
1440 |
0.82 |
Binding ≤ 10μM
|
|
CAH6-3-E |
Carbonic Anhydrase VI (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
1410 |
0.82 |
Binding ≤ 10μM
|
|
CAH7-3-E |
Carbonic Anhydrase VII (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
1230 |
0.83 |
Binding ≤ 10μM
|
|
CAH9-4-E |
Carbonic Anhydrase IX (cluster #4 Of 11), Eukaryotic |
Eukaryotes |
1370 |
0.82 |
Binding ≤ 10μM
|
|
Z80712-6-O |
T47D (Breast Carcinoma Cells) (cluster #6 Of 7), Other |
Other |
4400 |
0.75 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
113 |
0.97 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.83 |
-0.71 |
-175.27 |
8 |
3 |
3 |
72 |
148.274 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.83 |
-1.09 |
-97.55 |
7 |
3 |
2 |
70 |
147.266 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.83 |
-2.05 |
-90.25 |
7 |
3 |
2 |
67 |
147.266 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
360 |
0.43 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
520 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
0.63 |
-7.58 |
2 |
4 |
0 |
68 |
278.311 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
360 |
0.43 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
520 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
0.61 |
-7.58 |
2 |
4 |
0 |
68 |
278.311 |
2 |
↓
|
|
|
|
Analogs
-
6668761
-
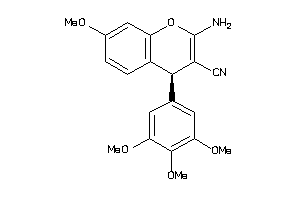
-
716753
-
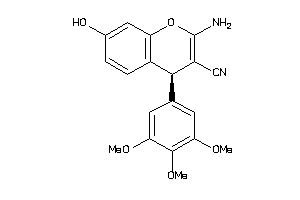
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
26 |
0.39 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
57 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
1.38 |
-13.01 |
2 |
7 |
0 |
95 |
368.389 |
5 |
↓
|
|
|
|
Analogs
-
716753
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
26 |
0.39 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
57 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
1.41 |
-12.6 |
2 |
7 |
0 |
95 |
368.389 |
5 |
↓
|
|
|
|
Analogs
-
5335560
-
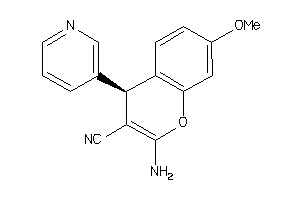
-
108024
-
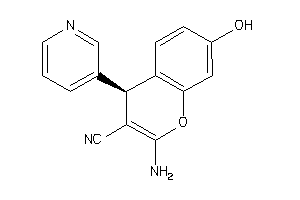
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
170 |
0.45 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
290 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
3.8 |
-9.65 |
2 |
5 |
0 |
81 |
279.299 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.54 |
4.08 |
-35.85 |
3 |
5 |
1 |
82 |
280.307 |
2 |
↓
|
|
|
|
Analogs
-
5335553
-
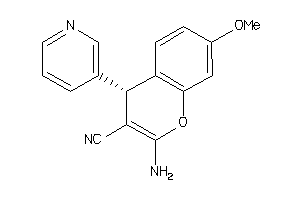
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
170 |
0.45 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
290 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
3.79 |
-9.67 |
2 |
5 |
0 |
81 |
279.299 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.54 |
4.07 |
-35.88 |
3 |
5 |
1 |
82 |
280.307 |
2 |
↓
|
|
|
|
Analogs
-
34044610
-
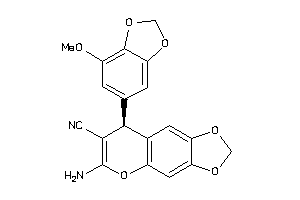
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
210 |
0.35 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
270 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
2.88 |
-15.04 |
2 |
8 |
0 |
105 |
366.329 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.29 |
3.21 |
-62.38 |
3 |
8 |
1 |
107 |
367.337 |
2 |
↓
|
|
|
|
Analogs
-
34044609
-
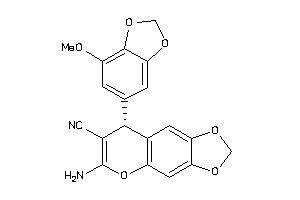
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
210 |
0.35 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
270 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
2.88 |
-14.92 |
2 |
8 |
0 |
105 |
366.329 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.29 |
3.23 |
-61.98 |
3 |
8 |
1 |
107 |
367.337 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
66 |
0.37 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
700 |
0.32 |
Functional ≤ 10μM |
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
58 |
0.38 |
Functional ≤ 10μM |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
19 |
0.40 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
43 |
0.38 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
9 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
6.62 |
-10.11 |
2 |
6 |
0 |
81 |
430.302 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
66 |
0.37 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
700 |
0.32 |
Functional ≤ 10μM |
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
58 |
0.38 |
Functional ≤ 10μM |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
19 |
0.40 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
43 |
0.38 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
9 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
6.63 |
-10.05 |
2 |
6 |
0 |
81 |
430.302 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
1000 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
-2.67 |
-9.78 |
1 |
5 |
0 |
56 |
361.426 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
158 |
0.41 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
112 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
7.2 |
-7.3 |
2 |
4 |
0 |
62 |
325.799 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
158 |
0.41 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
112 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
7.22 |
-7.68 |
2 |
4 |
0 |
62 |
325.799 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
270 |
0.44 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
200 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
-0.41 |
-7.83 |
2 |
4 |
0 |
62 |
297.383 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
270 |
0.44 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
200 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
-0.25 |
-7.76 |
2 |
4 |
0 |
62 |
297.383 |
2 |
↓
|
|
|
|
Analogs
-
8783644
-
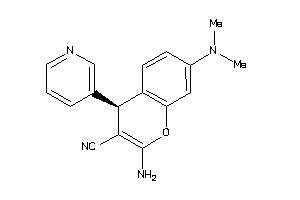
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
123 |
0.44 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
68 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
-0.2 |
-9.2 |
2 |
5 |
0 |
75 |
292.342 |
2 |
↓
|
|
|
|
Analogs
-
8783404
-
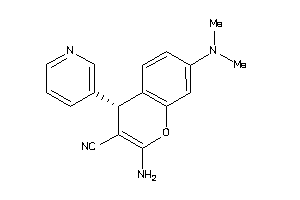
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
123 |
0.44 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
68 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
-0.38 |
-9.32 |
2 |
5 |
0 |
75 |
292.342 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
129 |
0.42 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
81 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
6.77 |
-7.98 |
2 |
4 |
0 |
62 |
309.344 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
129 |
0.42 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
81 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
6.78 |
-7.91 |
2 |
4 |
0 |
62 |
309.344 |
2 |
↓
|
|
|
|
Analogs
-
377693
-
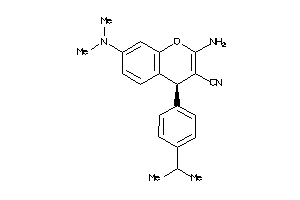
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
169 |
0.43 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
118 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
6.7 |
-7.41 |
2 |
4 |
0 |
62 |
291.354 |
2 |
↓
|
|
|
|
Analogs
-
377693
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
169 |
0.43 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
118 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
6.69 |
-7.38 |
2 |
4 |
0 |
62 |
291.354 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
27 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
1.53 |
-13.02 |
2 |
6 |
0 |
86 |
388.423 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
27 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
1.53 |
-12.77 |
2 |
6 |
0 |
86 |
388.423 |
4 |
↓
|
|
|
|
Analogs
-
33965412
-
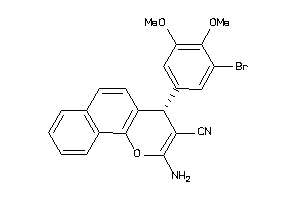
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
73 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
8.54 |
-11 |
2 |
5 |
0 |
78 |
437.293 |
3 |
↓
|
|
|
|
Analogs
-
33965411
-
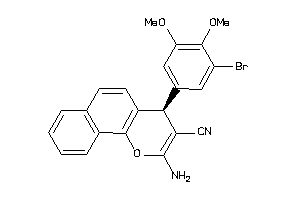
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
73 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
8.54 |
-10.72 |
2 |
5 |
0 |
78 |
437.293 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
110 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
4.54 |
-11.56 |
3 |
5 |
0 |
88 |
288.31 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
110 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
4.55 |
-11.57 |
3 |
5 |
0 |
88 |
288.31 |
1 |
↓
|
|
|
|
Analogs
-
1532612
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 36 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH14-2-E |
Carbonic Anhydrase XIV (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
860 |
0.61 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
10 |
0.80 |
Binding ≤ 10μM
|
|
CAH5A-4-E |
Carbonic Anhydrase VA (cluster #4 Of 10), Eukaryotic |
Eukaryotes |
840 |
0.61 |
Binding ≤ 10μM
|
|
CAH5B-5-E |
Carbonic Anhydrase VB (cluster #5 Of 9), Eukaryotic |
Eukaryotes |
830 |
0.61 |
Binding ≤ 10μM
|
|
CAH6-3-E |
Carbonic Anhydrase VI (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
990 |
0.60 |
Binding ≤ 10μM
|
|
CAH7-3-E |
Carbonic Anhydrase VII (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
710 |
0.61 |
Binding ≤ 10μM
|
|
CASP2-3-E |
Caspase-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.51 |
Binding ≤ 10μM
|
|
Z100250-2-O |
Calf Thymus DNA (cluster #2 Of 2), Other |
Other |
4300 |
0.54 |
Binding ≤ 10μM
|
|
Z104302-6-O |
Glutamate NMDA Receptor (cluster #6 Of 7), Other |
Other |
5200 |
0.53 |
Binding ≤ 10μM
|
|
Z80193-7-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #7 Of 12), Other |
Other |
390 |
0.64 |
Functional ≤ 10μM
|
|
Z80608-2-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #2 Of 4), Other |
Other |
200 |
0.67 |
Functional ≤ 10μM
|
|
Z80712-6-O |
T47D (Breast Carcinoma Cells) (cluster #6 Of 7), Other |
Other |
920 |
0.60 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
280 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.13 |
0.14 |
-262.68 |
10 |
4 |
4 |
88 |
206.378 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.13 |
-0.62 |
-97.52 |
8 |
4 |
2 |
85 |
204.362 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.13 |
-1.59 |
-88.23 |
8 |
4 |
2 |
82 |
204.362 |
11 |
↓
|
|