|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
22012961
-
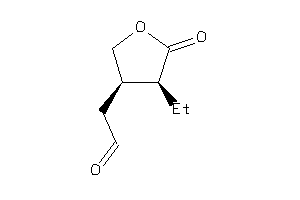
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.60 |
4.32 |
-11.21 |
0 |
3 |
0 |
43 |
156.181 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.60 |
4.32 |
-12.66 |
0 |
3 |
0 |
43 |
156.181 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.60 |
4.19 |
-14.09 |
0 |
3 |
0 |
43 |
156.181 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
11.57 |
-11.63 |
1 |
5 |
0 |
59 |
394.515 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.66 |
-2.48 |
-64.16 |
5 |
12 |
0 |
182 |
415.428 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.21 |
-3.43 |
-68.08 |
4 |
12 |
-1 |
185 |
414.42 |
4 |
↓
|
|
|
|
Analogs
-
1567835
-
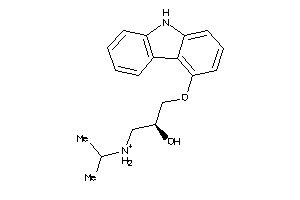
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
2.1 |
-51.06 |
5 |
4 |
1 |
73 |
257.313 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.64 |
1.72 |
-10.39 |
4 |
4 |
0 |
71 |
256.305 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.47 |
-1.91 |
-37.01 |
2 |
3 |
-1 |
44 |
152.22 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.47 |
-1.91 |
-37.02 |
2 |
3 |
-1 |
44 |
152.22 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
8.03 |
-5.48 |
0 |
2 |
0 |
10 |
188.274 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 3), Other |
Other |
3300 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.52 |
1.94 |
-67.91 |
4 |
6 |
-1 |
128 |
258.19 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
10.77 |
-2.96 |
0 |
0 |
0 |
0 |
210.32 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
11.36 |
-41.1 |
0 |
2 |
0 |
40 |
260.358 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.54 |
9.56 |
-5.26 |
0 |
4 |
0 |
55 |
318.543 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.71 |
-8.23 |
-495.03 |
4 |
15 |
-5 |
275 |
415.053 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.71 |
-7.09 |
-636.76 |
3 |
15 |
-6 |
278 |
414.045 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.71 |
-8.24 |
-452.98 |
4 |
15 |
-5 |
275 |
415.053 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
-0.28 |
-0.62 |
-33.47 |
3 |
2 |
1 |
29 |
101.173 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
-0.28 |
-0.61 |
-32.58 |
3 |
2 |
1 |
29 |
101.173 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
370 |
0.26 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
370 |
0.26 |
Binding ≤ 10μM
|
|
KCC2D-1-E |
CaM Kinase II Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8400 |
0.21 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
370 |
0.26 |
Binding ≤ 10μM
|
|
MYLK-1-E |
Myosin Light Chain Kinase, Smooth Muscle (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9800 |
0.21 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCC2A_MOUSE |
P11798
|
CaM Kinase II Alpha, Mouse |
370 |
0.26 |
Binding ≤ 1μM
|
|
KCC2B_MOUSE |
P28652
|
CaM Kinase II Beta, Mouse |
370 |
0.26 |
Binding ≤ 1μM
|
|
KCC2G_MOUSE |
Q923T9
|
CaM Kinase II Gamma, Mouse |
370 |
0.26 |
Binding ≤ 1μM
|
|
KCC2A_MOUSE |
P11798
|
CaM Kinase II Alpha, Mouse |
370 |
0.26 |
Binding ≤ 10μM
|
|
KCC2A_RAT |
P11275
|
CaM Kinase II Alpha, Rat |
8400 |
0.21 |
Binding ≤ 10μM
|
|
KCC2B_MOUSE |
P28652
|
CaM Kinase II Beta, Mouse |
370 |
0.26 |
Binding ≤ 10μM
|
|
KCC2B_RAT |
P08413
|
CaM Kinase II Beta, Rat |
8400 |
0.21 |
Binding ≤ 10μM
|
|
KCC2D_RAT |
P15791
|
CaM Kinase II Delta, Rat |
8400 |
0.21 |
Binding ≤ 10μM
|
|
KCC2G_MOUSE |
Q923T9
|
CaM Kinase II Gamma, Mouse |
370 |
0.26 |
Binding ≤ 10μM
|
|
KCC2G_RAT |
P11730
|
CaM Kinase II Gamma, Rat |
8400 |
0.21 |
Binding ≤ 10μM
|
|
MYLK_CHICK |
P11799
|
Myosin Light Chain Kinase, Smooth Muscle, Chick |
9800 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.58 |
7.11 |
-41.51 |
0 |
7 |
0 |
82 |
458.561 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
6.87 |
-9.82 |
2 |
5 |
0 |
85 |
280.283 |
3 |
↓
|
|
|
|
Analogs
-
4862976
-
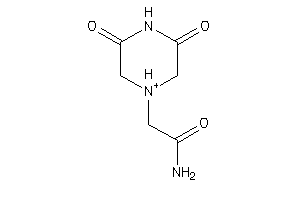
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.04 |
-3.37 |
-70.63 |
3 |
8 |
1 |
100 |
255.254 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.04 |
-5.69 |
-16.75 |
2 |
8 |
0 |
99 |
254.246 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.04 |
-1.07 |
-168.55 |
4 |
8 |
2 |
101 |
256.262 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
-2.99 |
-14.16 |
2 |
8 |
0 |
125 |
237.219 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.96 |
-9.65 |
-18.53 |
7 |
13 |
0 |
213 |
454.425 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
1.53 |
2.84 |
-56.8 |
4 |
11 |
-1 |
189 |
475.437 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
12.35 |
-92.9 |
0 |
0 |
2 |
0 |
308.386 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.13 |
11.59 |
-4.7 |
0 |
0 |
0 |
0 |
306.37 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.13 |
11.9 |
-29.86 |
0 |
0 |
1 |
0 |
307.378 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.56 |
2.73 |
-10.49 |
3 |
4 |
0 |
63 |
186.284 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.39 |
2.82 |
-5.56 |
3 |
4 |
0 |
63 |
186.284 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.56 |
2.93 |
-26.9 |
4 |
4 |
1 |
64 |
187.292 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
10.81 |
-126.13 |
1 |
9 |
-2 |
150 |
506.238 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
9.72 |
-40.78 |
2 |
9 |
-1 |
148 |
507.246 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
3.53 |
-53.06 |
3 |
3 |
1 |
49 |
241.261 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.99 |
3.84 |
-43.83 |
3 |
3 |
1 |
49 |
241.261 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.77 |
-4.96 |
-33.74 |
2 |
7 |
-1 |
110 |
152.093 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
-0.77 |
-5.01 |
-31.2 |
2 |
7 |
-1 |
110 |
152.093 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.77 |
-6.06 |
-35.4 |
2 |
7 |
-1 |
110 |
152.093 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.21 |
8.93 |
-46.3 |
1 |
3 |
-1 |
60 |
265.373 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.21 |
6.96 |
-7.32 |
2 |
3 |
0 |
58 |
266.381 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.18 |
3.06 |
-4.59 |
0 |
4 |
0 |
52 |
176.204 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.93 |
3.63 |
-40.54 |
1 |
4 |
-1 |
54 |
177.212 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.93 |
3.73 |
-28.06 |
2 |
4 |
0 |
56 |
178.22 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.79 |
-2 |
-4.45 |
4 |
4 |
0 |
70 |
182.223 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.49 |
-2.6 |
-135.27 |
4 |
12 |
-2 |
192 |
331.181 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.49 |
-3.75 |
-48.16 |
5 |
12 |
-1 |
189 |
332.189 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
11.47 |
-59.29 |
0 |
8 |
-1 |
112 |
410.413 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.40 |
11.82 |
-77.41 |
1 |
8 |
0 |
114 |
411.421 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.04 |
3.75 |
-10.4 |
0 |
2 |
0 |
26 |
132.546 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
1.78 |
-34.44 |
4 |
4 |
1 |
73 |
112.112 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.24 |
1.33 |
-16.12 |
3 |
4 |
0 |
72 |
111.104 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.44 |
13.64 |
-43.32 |
1 |
4 |
1 |
41 |
425.552 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.23 |
Functional ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Functional ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
5620 |
0.20 |
Functional ≤ 10μM
|
|
Z80175-1-O |
IMR-32 (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
1600 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
14.86 |
-19.33 |
1 |
9 |
0 |
120 |
492.528 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.23 |
Functional ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Functional ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
5620 |
0.20 |
Functional ≤ 10μM
|
|
Z80175-1-O |
IMR-32 (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
1600 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
14.87 |
-18.72 |
1 |
9 |
0 |
120 |
492.528 |
12 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
5.94 |
-208.03 |
1 |
9 |
-3 |
151 |
358.263 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.29 |
4.8 |
-103.89 |
2 |
9 |
-2 |
148 |
359.271 |
8 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.28 |
4.39 |
-45.69 |
3 |
1 |
1 |
28 |
148.229 |
1 |
↓
|
|