|
|
Analogs
-
12417074
-
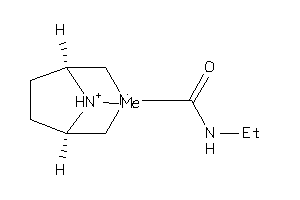
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.26 |
0.26 |
-44.01 |
1 |
4 |
1 |
28 |
226.344 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
5 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
10.85 |
-52.17 |
2 |
8 |
1 |
84 |
444.943 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
9 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
12.86 |
-47.06 |
1 |
8 |
1 |
72 |
458.97 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.48 |
10.54 |
-12.14 |
0 |
8 |
0 |
71 |
457.962 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
12.81 |
-49.88 |
2 |
7 |
1 |
71 |
489.603 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.22 |
10.49 |
-13.6 |
1 |
7 |
0 |
70 |
488.595 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.22 |
13.27 |
-87.02 |
3 |
7 |
2 |
72 |
490.611 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.29 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
1900 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
14.96 |
-50 |
1 |
7 |
1 |
60 |
503.63 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.76 |
12.65 |
-13.9 |
0 |
7 |
0 |
59 |
502.622 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.76 |
15.42 |
-87.55 |
2 |
7 |
2 |
61 |
504.638 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
170 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.11 |
13.37 |
-49.03 |
2 |
7 |
1 |
71 |
524.048 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.11 |
11.82 |
-55.15 |
0 |
7 |
-1 |
73 |
522.032 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.11 |
11.06 |
-12.48 |
1 |
7 |
0 |
70 |
523.04 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
6.96 |
-48.32 |
3 |
5 |
1 |
62 |
341.41 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
6.31 |
-44.5 |
3 |
6 |
1 |
75 |
372.518 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
5.53 |
-45.8 |
3 |
7 |
1 |
80 |
341.439 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
7.03 |
-42.33 |
2 |
4 |
1 |
46 |
247.366 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.64 |
5.75 |
-4.16 |
1 |
4 |
0 |
41 |
246.358 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.64 |
7.47 |
-84.38 |
3 |
4 |
2 |
47 |
248.374 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
6.52 |
-49.9 |
2 |
6 |
1 |
72 |
350.345 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.89 |
5.44 |
-10.99 |
1 |
6 |
0 |
68 |
349.337 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
4.15 |
-93.49 |
4 |
7 |
2 |
84 |
306.414 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.24 |
2.88 |
-50.09 |
3 |
7 |
1 |
80 |
305.406 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
4.05 |
-93.78 |
4 |
7 |
2 |
84 |
306.414 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.24 |
2.78 |
-50.32 |
3 |
7 |
1 |
80 |
305.406 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.73 |
3.14 |
-45.95 |
2 |
6 |
1 |
64 |
344.435 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.73 |
2.08 |
-8.96 |
1 |
6 |
0 |
60 |
343.427 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-1-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
5.24 |
-47.11 |
2 |
4 |
1 |
54 |
309.455 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.98 |
3.96 |
-7.48 |
1 |
4 |
0 |
49 |
308.447 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
5.87 |
-46.19 |
4 |
6 |
1 |
78 |
362.457 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
8.1 |
-56.77 |
2 |
6 |
1 |
63 |
387.46 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.56 |
6.83 |
-13.86 |
1 |
6 |
0 |
59 |
386.452 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
4.43 |
-42.93 |
2 |
4 |
1 |
46 |
318.828 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.74 |
3.37 |
-6.16 |
1 |
4 |
0 |
41 |
317.82 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
7.06 |
-52.94 |
3 |
6 |
1 |
75 |
352.462 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.92 |
5.97 |
-10.99 |
2 |
6 |
0 |
70 |
351.454 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.92 |
7.4 |
-89.63 |
4 |
6 |
2 |
76 |
353.47 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.59 |
6.19 |
-51.67 |
2 |
6 |
1 |
80 |
333.437 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.59 |
4.91 |
-15.85 |
1 |
6 |
0 |
75 |
332.429 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
5.09 |
-43.71 |
2 |
5 |
1 |
55 |
328.436 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.47 |
4.02 |
-9.46 |
1 |
5 |
0 |
51 |
327.428 |
5 |
↓
|
|