|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
10.81 |
-91.01 |
3 |
3 |
2 |
34 |
327.859 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.66 |
1.24 |
-17.31 |
2 |
7 |
0 |
85 |
443.503 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.54 |
15.55 |
-14.71 |
2 |
8 |
0 |
106 |
520.589 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.71 |
15.99 |
-42.57 |
3 |
8 |
1 |
111 |
521.597 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
11.8 |
-47.1 |
2 |
6 |
0 |
76 |
402.495 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.75 |
10.25 |
-47.84 |
1 |
6 |
-1 |
75 |
401.487 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.75 |
11.79 |
-80.85 |
3 |
6 |
1 |
77 |
403.503 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.26 |
12.03 |
-45.64 |
2 |
6 |
0 |
76 |
418.95 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.26 |
10.47 |
-47.46 |
1 |
6 |
-1 |
75 |
417.942 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.26 |
11.96 |
-79.97 |
3 |
6 |
1 |
77 |
419.958 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
12.2 |
-53.13 |
2 |
6 |
0 |
76 |
416.934 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.55 |
10.93 |
-50.49 |
1 |
6 |
-1 |
75 |
415.926 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.55 |
12.11 |
-88.77 |
3 |
6 |
1 |
77 |
417.942 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
12.05 |
-53.21 |
2 |
6 |
0 |
76 |
398.532 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.03 |
10.24 |
-53.25 |
1 |
6 |
0 |
75 |
397.524 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.03 |
11.97 |
-60.53 |
3 |
6 |
0 |
77 |
399.54 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.64 |
11.72 |
-48.7 |
2 |
7 |
0 |
86 |
414.531 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.64 |
10.22 |
-48.41 |
1 |
7 |
-1 |
85 |
413.523 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.64 |
11.68 |
-81.62 |
3 |
7 |
1 |
86 |
415.539 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
9.66 |
-48.93 |
3 |
7 |
0 |
97 |
400.504 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.36 |
7.99 |
-50.29 |
2 |
7 |
-1 |
96 |
399.496 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.36 |
9.45 |
-82.71 |
4 |
7 |
1 |
97 |
401.512 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
12.53 |
-49.36 |
2 |
6 |
0 |
76 |
398.532 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.08 |
11.05 |
-49.07 |
1 |
6 |
-1 |
75 |
397.524 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.08 |
12.33 |
-82.25 |
3 |
6 |
1 |
77 |
399.54 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.46 |
12.68 |
-54.47 |
2 |
6 |
0 |
76 |
412.559 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.46 |
11.29 |
-42.73 |
1 |
6 |
0 |
75 |
411.551 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.46 |
12.59 |
-61.17 |
3 |
6 |
0 |
77 |
413.567 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
9.03 |
-50.28 |
3 |
7 |
0 |
97 |
400.504 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.15 |
7.43 |
-48.4 |
2 |
7 |
-1 |
96 |
399.496 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.15 |
8.93 |
-85.12 |
4 |
7 |
1 |
97 |
401.512 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.67 |
2.77 |
-10.45 |
2 |
4 |
0 |
61 |
201.229 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
8.02 |
-19.03 |
2 |
10 |
0 |
131 |
512.588 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.24 |
6.64 |
-55.3 |
1 |
10 |
-1 |
137 |
511.58 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
10.78 |
-21.57 |
2 |
7 |
0 |
100 |
417.465 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.13 |
10.38 |
-62.59 |
1 |
7 |
-1 |
99 |
416.457 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
5.57 |
-7.08 |
3 |
5 |
0 |
81 |
259.309 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.34 |
5.7 |
-6.28 |
3 |
5 |
0 |
82 |
259.309 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.34 |
5.7 |
-6.59 |
3 |
5 |
0 |
82 |
259.309 |
4 |
↓
|
|
|
|
Analogs
-
6317908
-
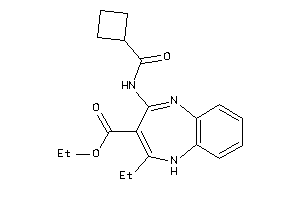
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
11 |
-10.82 |
2 |
9 |
0 |
121 |
466.538 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
3.26 |
10.87 |
-18.4 |
2 |
9 |
0 |
121 |
466.538 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
3.26 |
10.81 |
-16.93 |
2 |
9 |
0 |
121 |
466.538 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.28 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7400 |
0.27 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
38 |
0.38 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
8700 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
4.79 |
-18.75 |
1 |
7 |
0 |
79 |
386.477 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.90 |
7.14 |
-51.07 |
2 |
7 |
1 |
80 |
387.485 |
3 |
↓
|
|
|
|
Analogs
-
8972252
-
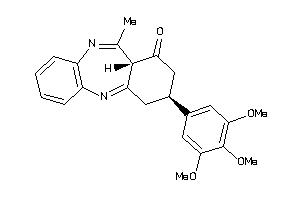
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
8.67 |
-16.6 |
1 |
6 |
0 |
73 |
392.455 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.99 |
8.75 |
-12.41 |
1 |
6 |
0 |
73 |
392.455 |
4 |
↓
|
|
|
|
Analogs
-
8972252
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
8.67 |
-16.85 |
1 |
6 |
0 |
73 |
392.455 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.99 |
8.75 |
-12.42 |
1 |
6 |
0 |
73 |
392.455 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
8.23 |
-14.07 |
1 |
5 |
0 |
64 |
362.429 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.92 |
8.33 |
-10 |
1 |
5 |
0 |
64 |
362.429 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
8.23 |
-13.12 |
1 |
5 |
0 |
64 |
362.429 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.92 |
8.33 |
-10.01 |
1 |
5 |
0 |
64 |
362.429 |
3 |
↓
|
|
|
|
Analogs
-
9058745
-
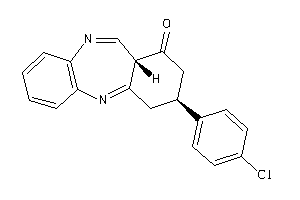
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
10.06 |
-11.96 |
1 |
3 |
0 |
46 |
336.822 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
4.94 |
8.7 |
-41.44 |
0 |
3 |
-1 |
49 |
335.814 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.03 |
10.16 |
-7.5 |
1 |
3 |
0 |
46 |
336.822 |
1 |
↓
|
|
|
|
Analogs
-
9058745
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
10.06 |
-11.85 |
1 |
3 |
0 |
46 |
336.822 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
4.94 |
8.7 |
-41.49 |
0 |
3 |
-1 |
49 |
335.814 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.03 |
10.16 |
-7.55 |
1 |
3 |
0 |
46 |
336.822 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
6.06 |
-51 |
1 |
7 |
-1 |
103 |
369.426 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.83 |
7.56 |
-14.4 |
2 |
7 |
0 |
97 |
370.434 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.30 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.30 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
300 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.69 |
9.69 |
-16.43 |
2 |
8 |
0 |
116 |
416.828 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.34 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.34 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
50 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
9.7 |
-15.45 |
2 |
8 |
0 |
116 |
416.828 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.37 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.37 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
20 |
0.37 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
5300 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
9.15 |
-14.88 |
3 |
8 |
0 |
119 |
404.817 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
46 |
0.37 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
46 |
0.37 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
46 |
0.37 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
1100 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
4.28 |
-14.49 |
3 |
8 |
0 |
107 |
416.894 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.37 |
4.32 |
-49.66 |
2 |
8 |
-1 |
109 |
415.886 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.37 |
4.46 |
-42.42 |
4 |
8 |
1 |
108 |
417.902 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.23 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.23 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.31 |
12.93 |
-15.16 |
2 |
7 |
0 |
103 |
443.894 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.31 |
13.1 |
-49.78 |
3 |
7 |
1 |
104 |
444.902 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
210 |
0.31 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
210 |
0.31 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
210 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.76 |
11.44 |
-14.75 |
2 |
7 |
0 |
103 |
415.84 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.76 |
11.64 |
-45.68 |
3 |
7 |
1 |
104 |
416.848 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.40 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.40 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
440 |
0.40 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
3600 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
7.68 |
-9.23 |
2 |
4 |
0 |
57 |
329.19 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.81 |
7.85 |
-35.43 |
3 |
4 |
1 |
59 |
330.198 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
258 |
0.34 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
258 |
0.34 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
258 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
4.66 |
-16.04 |
4 |
9 |
0 |
146 |
382.767 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK2_HUMAN |
P24941
|
Cyclin-dependent Kinase 2, Human |
500 |
0.37 |
Binding ≤ 1μM
|
|
CCNE1_HUMAN |
P24864
|
G1/S-specific Cyclin E1, Human |
70 |
0.42 |
Binding ≤ 1μM
|
|
CCNE2_HUMAN |
O96020
|
G1/S-specific Cyclin E2, Human |
70 |
0.42 |
Binding ≤ 1μM
|
|
CCNB1_HUMAN |
P14635
|
G2/mitotic-specific Cyclin B1, Human |
500 |
0.37 |
Binding ≤ 1μM
|
|
CCNB2_HUMAN |
O95067
|
G2/mitotic-specific Cyclin B2, Human |
500 |
0.37 |
Binding ≤ 1μM
|
|
CCNB3_HUMAN |
Q8WWL7
|
G2/mitotic-specific Cyclin B3, Human |
500 |
0.37 |
Binding ≤ 1μM
|
|
CDK2_HUMAN |
P24941
|
Cyclin-dependent Kinase 2, Human |
10000 |
0.29 |
Binding ≤ 10μM
|
|
CCND1_HUMAN |
P24385
|
G1/S-specific Cyclin D1, Human |
10000 |
0.29 |
Binding ≤ 10μM
|
|
CCNE1_HUMAN |
P24864
|
G1/S-specific Cyclin E1, Human |
70 |
0.42 |
Binding ≤ 10μM
|
|
CCNE2_HUMAN |
O96020
|
G1/S-specific Cyclin E2, Human |
70 |
0.42 |
Binding ≤ 10μM
|
|
CCNB1_HUMAN |
P14635
|
G2/mitotic-specific Cyclin B1, Human |
500 |
0.37 |
Binding ≤ 10μM
|
|
CCNB2_HUMAN |
O95067
|
G2/mitotic-specific Cyclin B2, Human |
500 |
0.37 |
Binding ≤ 10μM
|
|
CCNB3_HUMAN |
Q8WWL7
|
G2/mitotic-specific Cyclin B3, Human |
500 |
0.37 |
Binding ≤ 10μM
|
|
Z80526 |
Z80526
|
SW480 (Colon Adenocarcinoma Cells) |
6000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
7.8 |
-15.92 |
2 |
7 |
0 |
103 |
339.742 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.09 |
7.98 |
-50.91 |
3 |
7 |
1 |
104 |
340.75 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.41 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.41 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.41 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
640 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.37 |
9.75 |
-14.1 |
2 |
7 |
0 |
103 |
381.823 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.37 |
9.92 |
-44.81 |
3 |
7 |
1 |
104 |
382.831 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
238 |
0.36 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
238 |
0.36 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
238 |
0.36 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
240 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.67 |
5.14 |
-14.89 |
3 |
8 |
0 |
123 |
369.768 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.67 |
5.33 |
-44.24 |
4 |
8 |
1 |
125 |
370.776 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.40 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.40 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
320 |
0.40 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
7000 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
6.45 |
-11.06 |
2 |
5 |
0 |
67 |
324.771 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.19 |
6.63 |
-35.26 |
3 |
5 |
1 |
68 |
325.779 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.45 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.45 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.45 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
210 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
3.63 |
-16.94 |
4 |
6 |
0 |
100 |
321.315 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.44 |
3.8 |
-43.39 |
5 |
6 |
1 |
102 |
322.323 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.41 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.41 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
24 |
0.41 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
200 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
3.94 |
-43.28 |
2 |
7 |
-1 |
106 |
386.844 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.37 |
3.88 |
-13.96 |
3 |
7 |
0 |
104 |
387.852 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.37 |
4.04 |
-43.52 |
4 |
7 |
1 |
105 |
388.86 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
303 |
0.41 |
Binding ≤ 10μM
|
|
CCND1-1-E |
G1/S-specific Cyclin D1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
382 |
0.41 |
Binding ≤ 10μM
|
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
88 |
0.45 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
88 |
0.45 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
382 |
0.41 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
88 |
0.45 |
Binding ≤ 10μM
|
|
CDK4-1-E |
Cyclin-dependent Kinase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
303 |
0.41 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
989 |
0.38 |
Binding ≤ 10μM
|
|
FAK1-1-E |
Focal Adhesion Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
150 |
0.43 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1840 |
0.36 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
34 |
0.48 |
Binding ≤ 10μM
|
|
Z80427-1-O |
RKO (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
310 |
0.41 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
500 |
0.40 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
1018 |
0.38 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
950 |
0.38 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1_HUMAN |
P78396
|
Cyclin A1, Human |
303 |
0.41 |
Binding ≤ 1μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
382 |
0.41 |
Binding ≤ 1μM
|
|
CDK2_HUMAN |
P24941
|
Cyclin-dependent Kinase 2, Human |
88 |
0.45 |
Binding ≤ 1μM
|
|
CDK4_HUMAN |
P11802
|
Cyclin-dependent Kinase 4, Human |
303 |
0.41 |
Binding ≤ 1μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
989 |
0.38 |
Binding ≤ 1μM
|
|
FAK1_HUMAN |
Q05397
|
Focal Adhesion Kinase 1, Human |
150 |
0.43 |
Binding ≤ 1μM
|
|
CCND1_HUMAN |
P24385
|
G1/S-specific Cyclin D1, Human |
382 |
0.41 |
Binding ≤ 1μM
|
|
CCNE1_HUMAN |
P24864
|
G1/S-specific Cyclin E1, Human |
88 |
0.45 |
Binding ≤ 1μM
|
|
CCNE2_HUMAN |
O96020
|
G1/S-specific Cyclin E2, Human |
88 |
0.45 |
Binding ≤ 1μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
34 |
0.48 |
Binding ≤ 1μM
|
|
CCNA1_HUMAN |
P78396
|
Cyclin A1, Human |
303 |
0.41 |
Binding ≤ 10μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
382 |
0.41 |
Binding ≤ 10μM
|
|
CDK2_HUMAN |
P24941
|
Cyclin-dependent Kinase 2, Human |
88 |
0.45 |
Binding ≤ 10μM
|
|
CDK4_HUMAN |
P11802
|
Cyclin-dependent Kinase 4, Human |
303 |
0.41 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
989 |
0.38 |
Binding ≤ 10μM
|
|
FAK1_HUMAN |
Q05397
|
Focal Adhesion Kinase 1, Human |
150 |
0.43 |
Binding ≤ 10μM
|
|
CCND1_HUMAN |
P24385
|
G1/S-specific Cyclin D1, Human |
382 |
0.41 |
Binding ≤ 10μM
|
|
CCNE1_HUMAN |
P24864
|
G1/S-specific Cyclin E1, Human |
88 |
0.45 |
Binding ≤ 10μM
|
|
CCNE2_HUMAN |
O96020
|
G1/S-specific Cyclin E2, Human |
88 |
0.45 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
1840 |
0.36 |
Binding ≤ 10μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
34 |
0.48 |
Binding ≤ 10μM
|
|
Z80928 |
Z80928
|
HCT-116 (Colon Carcinoma Cells) |
1018 |
0.38 |
Functional ≤ 10μM
|
|
Z81245 |
Z81245
|
MDA-MB-435 (Breast Carcinoma Cells) |
950 |
0.38 |
Functional ≤ 10μM
|
|
Z80427 |
Z80427
|
RKO (Colon Carcinoma) |
310 |
0.41 |
Functional ≤ 10μM
|
|
Z80526 |
Z80526
|
SW480 (Colon Adenocarcinoma Cells) |
500 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
5.04 |
-11 |
4 |
5 |
0 |
83 |
309.76 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.21 |
5.22 |
-33.13 |
5 |
5 |
1 |
85 |
310.768 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
213 |
0.37 |
Binding ≤ 10μM
|
|
CCND1-1-E |
G1/S-specific Cyclin D1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
352 |
0.36 |
Binding ≤ 10μM
|
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
38 |
0.42 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
38 |
0.42 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
352 |
0.36 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
38 |
0.42 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
213 |
0.37 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
726 |
0.34 |
Binding ≤ 10μM
|
|
FAK1-1-E |
Focal Adhesion Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
180 |
0.38 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
930 |
0.34 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.42 |
Binding ≤ 10μM
|
|
Z80427-1-O |
RKO (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
793 |
0.34 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
700 |
0.34 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
460 |
0.35 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
794 |
0.34 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1_HUMAN |
P78396
|
Cyclin A1, Human |
213 |
0.37 |
Binding ≤ 1μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
352 |
0.36 |
Binding ≤ 1μM
|
|
CDK2_HUMAN |
P24941
|
Cyclin-dependent Kinase 2, Human |
38 |
0.42 |
Binding ≤ 1μM
|
|
CDK4_HUMAN |
P11802
|
Cyclin-dependent Kinase 4, Human |
213 |
0.37 |
Binding ≤ 1μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
726 |
0.34 |
Binding ≤ 1μM
|
|
FAK1_HUMAN |
Q05397
|
Focal Adhesion Kinase 1, Human |
180 |
0.38 |
Binding ≤ 1μM
|
|
CCND1_HUMAN |
P24385
|
G1/S-specific Cyclin D1, Human |
352 |
0.36 |
Binding ≤ 1μM
|
|
CCNE1_HUMAN |
P24864
|
G1/S-specific Cyclin E1, Human |
38 |
0.42 |
Binding ≤ 1μM
|
|
CCNE2_HUMAN |
O96020
|
G1/S-specific Cyclin E2, Human |
38 |
0.42 |
Binding ≤ 1μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
930 |
0.34 |
Binding ≤ 1μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
33 |
0.42 |
Binding ≤ 1μM
|
|
CCNA1_HUMAN |
P78396
|
Cyclin A1, Human |
213 |
0.37 |
Binding ≤ 10μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
352 |
0.36 |
Binding ≤ 10μM
|
|
CDK2_HUMAN |
P24941
|
Cyclin-dependent Kinase 2, Human |
38 |
0.42 |
Binding ≤ 10μM
|
|
CDK4_HUMAN |
P11802
|
Cyclin-dependent Kinase 4, Human |
213 |
0.37 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
726 |
0.34 |
Binding ≤ 10μM
|
|
FAK1_HUMAN |
Q05397
|
Focal Adhesion Kinase 1, Human |
180 |
0.38 |
Binding ≤ 10μM
|
|
CCND1_HUMAN |
P24385
|
G1/S-specific Cyclin D1, Human |
352 |
0.36 |
Binding ≤ 10μM
|
|
CCNE1_HUMAN |
P24864
|
G1/S-specific Cyclin E1, Human |
38 |
0.42 |
Binding ≤ 10μM
|
|
CCNE2_HUMAN |
O96020
|
G1/S-specific Cyclin E2, Human |
38 |
0.42 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
930 |
0.34 |
Binding ≤ 10μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
33 |
0.42 |
Binding ≤ 10μM
|
|
Z80928 |
Z80928
|
HCT-116 (Colon Carcinoma Cells) |
460 |
0.35 |
Functional ≤ 10μM
|
|
Z81245 |
Z81245
|
MDA-MB-435 (Breast Carcinoma Cells) |
794 |
0.34 |
Functional ≤ 10μM
|
|
Z80427 |
Z80427
|
RKO (Colon Carcinoma) |
793 |
0.34 |
Functional ≤ 10μM
|
|
Z80526 |
Z80526
|
SW480 (Colon Adenocarcinoma Cells) |
700 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
6.07 |
-15.91 |
3 |
6 |
0 |
86 |
351.797 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.35 |
6.25 |
-39.49 |
4 |
6 |
1 |
88 |
352.805 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
115 |
0.46 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
115 |
0.46 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
115 |
0.46 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
1500 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
7.34 |
-9.78 |
2 |
4 |
0 |
57 |
274.327 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.75 |
7.5 |
-29.07 |
3 |
4 |
1 |
59 |
275.335 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
105 |
0.44 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
105 |
0.44 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
105 |
0.44 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
3800 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
7.72 |
-10.43 |
2 |
4 |
0 |
57 |
308.772 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.38 |
7.88 |
-29.73 |
3 |
4 |
1 |
59 |
309.78 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
232 |
0.33 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
232 |
0.33 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
232 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.23 |
8.79 |
-13.53 |
2 |
7 |
0 |
103 |
407.739 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
93 |
0.45 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
93 |
0.45 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
93 |
0.45 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
2200 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
7.42 |
-8.53 |
2 |
4 |
0 |
57 |
292.317 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.92 |
7.57 |
-31.81 |
3 |
4 |
1 |
59 |
293.325 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.45 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.45 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
40 |
0.45 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
700 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
7.46 |
-10.62 |
2 |
4 |
0 |
57 |
310.307 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.03 |
7.63 |
-32.95 |
3 |
4 |
1 |
59 |
311.315 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
490 |
0.42 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
490 |
0.42 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
490 |
0.42 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
5500 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
7.32 |
-8.48 |
2 |
4 |
0 |
57 |
294.745 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.18 |
7.48 |
-34.76 |
3 |
4 |
1 |
59 |
295.753 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
64 |
0.46 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
64 |
0.46 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
64 |
0.46 |
Binding ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
400 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.87 |
7.39 |
-12.06 |
2 |
4 |
0 |
57 |
292.317 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.87 |
7.55 |
-30.52 |
3 |
4 |
1 |
59 |
293.325 |
1 |
↓
|
|
|
|
|
|
|
|