|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4S,5R,6R)-3,4,5-trihydroxy-6-[3-[4-[(8S,11R,13S,14S,17S)-17-hydroxy-13-methyl-3-oxo-17-prop-1
(2R,3S,4S,5R,6R)-3,4,5-trihydrox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-1-E |
Glucocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.17 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
550 |
0.19 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
10.01 |
-74.76 |
4 |
9 |
0 |
151 |
632.774 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.92 |
10.45 |
-67.51 |
5 |
9 |
0 |
152 |
633.782 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4S,5R,6S)-3,4,5-trihydroxy-6-[3-[4-[(8S,11R,13S,14S,17S)-17-hydroxy-13-methyl-3-oxo-17-prop-1
(2R,3S,4S,5R,6S)-3,4,5-trihydrox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-1-E |
Glucocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.17 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
550 |
0.19 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
9.72 |
-60.16 |
4 |
9 |
-1 |
151 |
632.774 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.92 |
10.15 |
-72.51 |
5 |
9 |
0 |
152 |
633.782 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,11R,13S,14S,17R)-17-hydroxy-13-methyl-11-[4-(methylamino)phenyl]-17-prop-1-ynyl-1,2,6,7,8,11,12,
(8R,11R,13S,14S,17R)-17-hydroxy-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.35 |
10.68 |
-10.5 |
2 |
3 |
0 |
49 |
415.577 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.35 |
11.23 |
-45.85 |
3 |
3 |
1 |
54 |
416.585 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[4-[(8R,11R,13S,14S,17R)-17-hydroxy-13-methyl-3-oxo-17-prop-1-ynyl-1,2,6,7,8,11,12,14,15,16-decahy
3-[4-[(8R,11R,13S,14S,17R)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
34 |
0.29 |
Binding ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
34 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.11 |
13.89 |
-53.41 |
1 |
5 |
-1 |
81 |
486.632 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.11 |
14.33 |
-54.08 |
2 |
5 |
0 |
82 |
487.64 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
6-[4-[(8R,11R,13S,14S,17R)-17-hydroxy-13-methyl-3-oxo-17-prop-1-ynyl-1,2,6,7,8,11,12,14,15,16-decahy
6-[4-[(8R,11R,13S,14S,17R)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.39 |
16.27 |
-56.03 |
1 |
5 |
-1 |
81 |
528.713 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.39 |
16.69 |
-70.84 |
2 |
5 |
0 |
82 |
529.721 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.39 |
14.71 |
-78.05 |
3 |
5 |
0 |
79 |
530.729 |
8 |
↓
|
|
|
|
Analogs
-
44352463
-
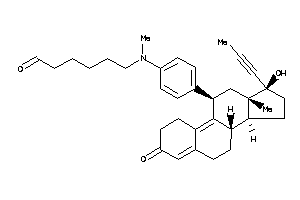
-
44359735
-
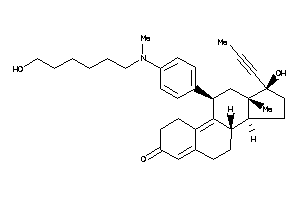
-
53276052
-
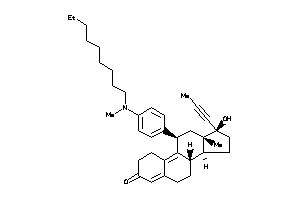
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,11R,13S,14S,17S)-11-[4-[hexyl(methyl)amino]phenyl]-17-hydroxy-13-methyl-17-prop-1-ynyl-1,2,6,7,8
(8S,11R,13S,14S,17S)-11-[4-[hexy…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.04 |
16.18 |
-9.58 |
1 |
3 |
0 |
41 |
499.739 |
7 |
↓
|
|
|
|
Analogs
-
968322
-
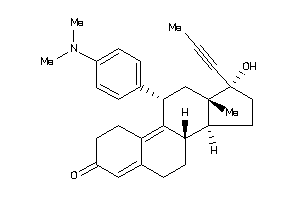
-
2100511
-
-
3814382
-
Draw
Identity
99%
90%
80%
70%
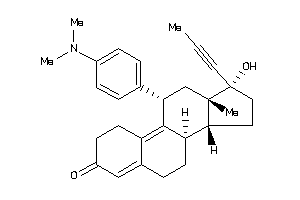
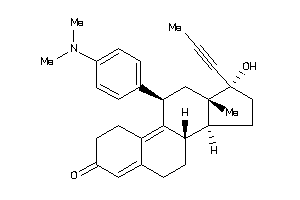
Popular Name:
(8S,11R,13R,14S,17R)-11-(4-dimethylaminophenyl)-17-hydroxy-13-methyl-17-prop-1-ynyl-1,2,6,7,8,11,12,
(8S,11R,13R,14S,17R)-11-(4-dimet…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.60 |
12.63 |
-10.36 |
1 |
3 |
0 |
41 |
429.604 |
2 |
↓
|
|