|
|
Analogs
-
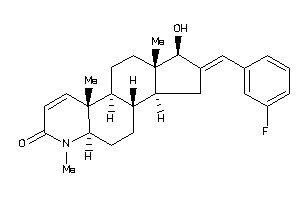
38412187
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)-2-[(3-fluorophenyl)methylene]-1-hydroxy-6,9a,11a-trimethyl-1,3,3a,3
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.35 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
29 |
0.35 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
10.05 |
-12.7 |
1 |
3 |
0 |
41 |
409.545 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aS)-2-[(3,5-difluorophenyl)methylene]-1-hydroxy-6,9a,11a-trimethyl-1,3,
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aS)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.21 |
10.08 |
-10.28 |
1 |
3 |
0 |
41 |
427.535 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)-2-benzylidene-1-hydroxy-6,9a,11a-trimethyl-1,3,3a,3b,4,5,5a,9b,10,1
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
116 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
225 |
0.32 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.95 |
9.98 |
-11.65 |
1 |
3 |
0 |
41 |
391.555 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)-2-ethylidene-1-hydroxy-6,9a,11a-trimethyl-1,3,3a,3b,4,5,5a,9b,10,11
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
35 |
0.43 |
Binding ≤ 10μM
|
|
KCNH2-4-E |
HERG (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
8050 |
0.30 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
17 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
7.24 |
-9.97 |
1 |
3 |
0 |
41 |
329.484 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)-1-hydroxy-6,9a,11a-trimethyl-2-(2-pyridylmethylene)-1,3,3a,3b,4,5,5
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
292 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
8.99 |
-13.52 |
1 |
4 |
0 |
53 |
392.543 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.60 |
9.15 |
-33.03 |
2 |
4 |
1 |
55 |
393.551 |
1 |
↓
|
|
|
|
Analogs
-
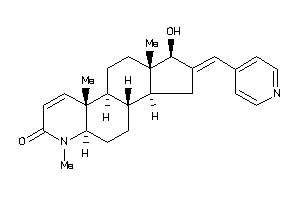
38411859
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)-1-hydroxy-6,9a,11a-trimethyl-2-(4-pyridylmethylene)-1,3,3a,3b,4,5,5
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
129 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-4-E |
HERG (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
320 |
0.31 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
138 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
8.12 |
-13.71 |
1 |
4 |
0 |
53 |
392.543 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.66 |
8.58 |
-36.32 |
2 |
4 |
1 |
55 |
393.551 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)-1-hydroxy-6,9a,11a-trimethyl-2-(3-pyridylmethylene)-1,3,3a,3b,4,5,5
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
62 |
0.35 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3600 |
0.26 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
62 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
8.11 |
-13.05 |
1 |
4 |
0 |
53 |
392.543 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.54 |
8.57 |
-37.12 |
2 |
4 |
1 |
55 |
393.551 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)-1-hydroxy-6,9a,11a-trimethyl-2-(pyrimidin-5-ylmethylene)-1,3,3a,3b,
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
59 |
0.35 |
Binding ≤ 10μM
|
|
KCNH2-4-E |
HERG (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
9400 |
0.24 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
17 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
7.89 |
-14.76 |
1 |
5 |
0 |
66 |
393.531 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aR,3bS,5aR,9aS,9bR,11aR)-2-[(2-cyclopropylpyrimidin-5-yl)methylene]-1-hydroxy-6,9a,11a-trime
(1R,2E,3aR,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
9.54 |
-14.59 |
1 |
5 |
0 |
66 |
433.596 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aS)-1-hydroxy-2-[(2-methoxypyrimidin-5-yl)methylene]-6,9a,11a-trimethyl
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aS)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
11 |
0.36 |
Binding ≤ 10μM
|
|
KCNH2-4-E |
HERG (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7900 |
0.23 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
4.61 |
-14.23 |
1 |
6 |
0 |
76 |
423.557 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aS)-1-hydroxy-6,9a,11a-trimethyl-2-[[2-(methylamino)pyrimidin-5-yl]meth
(1R,2E,3aS,3bS,5aR,9aS,9bR,11aS)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
31 |
0.34 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
8.42 |
-13 |
2 |
6 |
0 |
78 |
422.573 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2E,3aR,3bS,5aR,9aS,9bR,11aR)-1-hydroxy-6,9a,11a-trimethyl-2-[[2-(2-pyridyl)pyrimidin-5-yl]methyl
(1R,2E,3aR,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-1-E |
Androgen Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
715 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
9.31 |
-19.13 |
1 |
6 |
0 |
79 |
470.617 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,2E,3aS,3bR,5aR,9aR,9bS,11aS)-1-hydroxy-6,9a,11a-trimethyl-2-[[2-(2-pyridyl)pyrimidin-5-yl]methyl
(1S,2E,3aS,3bR,5aR,9aR,9bS,11aS)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
9.44 |
-18.95 |
1 |
6 |
0 |
79 |
470.617 |
2 |
↓
|
|