|
|
Analogs
-
29230271
-
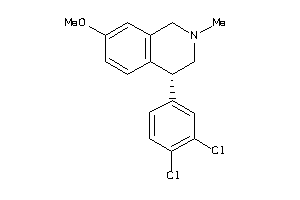
-
29230278
-
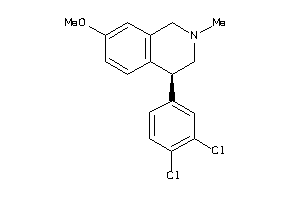
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
1.75 |
-45.69 |
1 |
2 |
1 |
13 |
323.243 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
565 |
0.49 |
Binding ≤ 1μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
Q8CFM9_RAT |
Q8CFM9
|
Adenylate Cyclase Type VII, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
3620 |
0.42 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
3620 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
5.6 |
-51.39 |
3 |
2 |
1 |
37 |
260.744 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.24 |
4.9 |
-43.88 |
1 |
2 |
-1 |
35 |
258.728 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.24 |
6.35 |
-72.44 |
2 |
2 |
0 |
40 |
259.736 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
565 |
0.49 |
Binding ≤ 1μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
Q8CFM9_RAT |
Q8CFM9
|
Adenylate Cyclase Type VII, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
1257 |
0.46 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
3620 |
0.42 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
3620 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
4.28 |
-5.37 |
2 |
2 |
0 |
32 |
259.736 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.24 |
4.94 |
-45.82 |
1 |
2 |
-1 |
35 |
258.728 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.24 |
6.33 |
-72.42 |
2 |
2 |
0 |
40 |
259.736 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
Q8CFM9_RAT |
Q8CFM9
|
Adenylate Cyclase Type VII, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
174 |
0.50 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
522 |
0.46 |
Binding ≤ 1μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
Q8CFM9_RAT |
Q8CFM9
|
Adenylate Cyclase Type VII, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
174 |
0.50 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
522 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
5.44 |
-6.6 |
1 |
2 |
0 |
23 |
273.763 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.48 |
6.22 |
-44.45 |
0 |
2 |
-1 |
26 |
272.755 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
Q8CFM9_RAT |
Q8CFM9
|
Adenylate Cyclase Type VII, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
205 |
0.49 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
174 |
0.50 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
522 |
0.46 |
Binding ≤ 1μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
Q8CFM9_RAT |
Q8CFM9
|
Adenylate Cyclase Type VII, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
205 |
0.49 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
174 |
0.50 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
522 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
5.44 |
-6.6 |
1 |
2 |
0 |
23 |
273.763 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.48 |
6.23 |
-44.47 |
0 |
2 |
-1 |
26 |
272.755 |
1 |
↓
|
|
|
|
Analogs
-
13892812
-
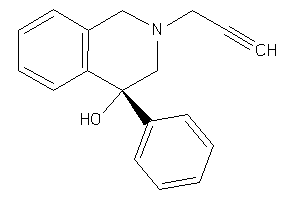
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
5.77 |
-6.35 |
1 |
2 |
0 |
23 |
263.34 |
2 |
↓
|
|