|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
5781610
-
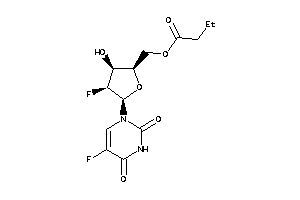
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.08 |
-2.26 |
-14.42 |
2 |
8 |
0 |
110 |
334.275 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
33815349
-
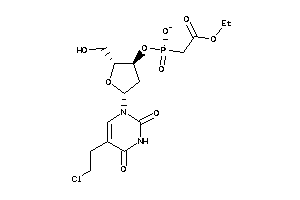
-
40374252
-
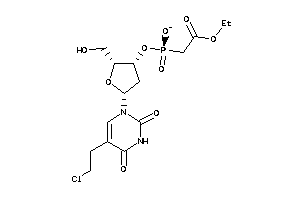
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.21 |
2.23 |
-54.27 |
2 |
11 |
-1 |
160 |
439.765 |
10 |
↓
|
|
|
|
Analogs
-
40374252
-

-
33815348
-
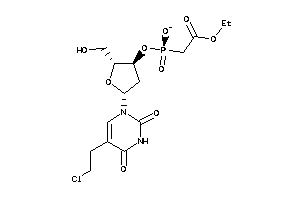
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.21 |
2.22 |
-54.97 |
2 |
11 |
-1 |
160 |
439.765 |
10 |
↓
|
|
|
|
Analogs
-
33815357
-
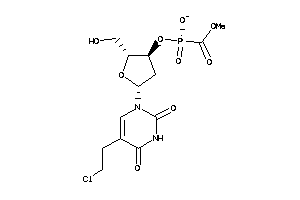
-
40374890
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.60 |
0.44 |
-51.51 |
2 |
11 |
-1 |
160 |
411.711 |
8 |
↓
|
|
|
|
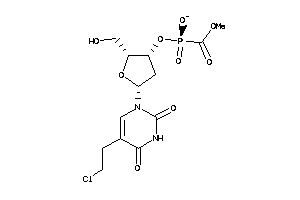
Analogs
-
40374890
-

-
33815356
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.60 |
0.39 |
-54.97 |
2 |
11 |
-1 |
160 |
411.711 |
8 |
↓
|
|
|
|
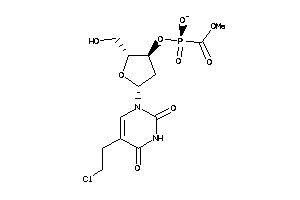
Analogs
-
33815359
-
-
40374752
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.22 |
1.48 |
-49.18 |
2 |
11 |
-1 |
160 |
425.738 |
9 |
↓
|
|
|
|
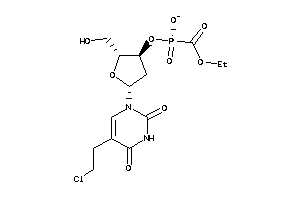
Analogs
-
40374752
-

-
33815358
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.22 |
1.3 |
-55.76 |
2 |
11 |
-1 |
160 |
425.738 |
9 |
↓
|
|
|
|
|
|
|
Analogs
-
33816393
-
-
33816394
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
3280 |
0.25 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547_9HIV1 |
Q72547
|
Human Immunodeficiency Virus Type 1 Reverse Transcriptase, 9hiv1 |
3280 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.88 |
-0.91 |
-374.1 |
2 |
16 |
-4 |
255 |
502.158 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.88 |
-2.07 |
-238.27 |
3 |
16 |
-3 |
253 |
503.166 |
8 |
↓
|
|
|
|
Analogs
-
8585026
-
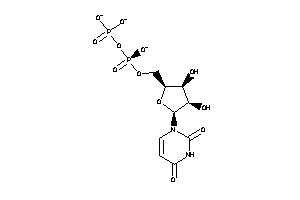
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.76 |
-4.62 |
-261.56 |
3 |
14 |
-3 |
226 |
401.137 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.76 |
-5.78 |
-142.15 |
4 |
14 |
-2 |
224 |
402.145 |
6 |
↓
|
|
|
|
Analogs
-
6524816
-
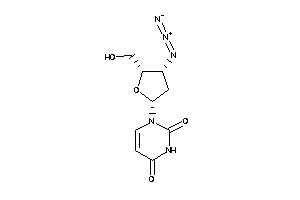
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.80 |
-3.69 |
-21.01 |
2 |
9 |
0 |
134 |
253.218 |
3 |
↓
|
|
|
|
Analogs
-
26154932
-
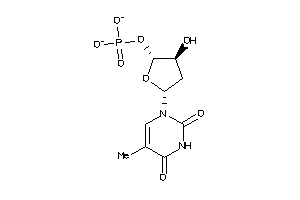
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
2700 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-2.13 |
-129.37 |
2 |
10 |
-2 |
157 |
306.167 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.11 |
-3.27 |
-43.28 |
3 |
10 |
-1 |
154 |
307.175 |
3 |
↓
|
|
|
|
Analogs
-
26154932
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
2700 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-1.51 |
-144.29 |
2 |
10 |
-2 |
157 |
306.167 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.11 |
-2.66 |
-55.26 |
3 |
10 |
-1 |
154 |
307.175 |
3 |
↓
|
|
|
|
Analogs
-
26154932
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
2700 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-2.86 |
-127.26 |
2 |
10 |
-2 |
157 |
306.167 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.11 |
-4.01 |
-41.97 |
3 |
10 |
-1 |
154 |
307.175 |
3 |
↓
|
|
|
|
Analogs
-
26154932
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2700 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
2700 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-2.21 |
-148.7 |
2 |
10 |
-2 |
157 |
306.167 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.11 |
-3.36 |
-59.9 |
3 |
10 |
-1 |
154 |
307.175 |
3 |
↓
|
|
|
|
Analogs
-
11990271
-
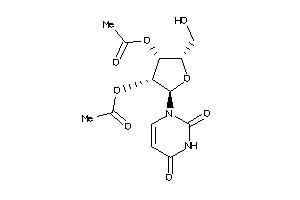
-
4367689
-
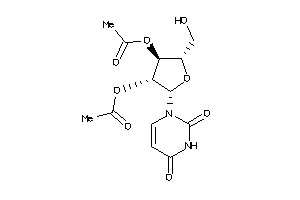
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.64 |
-3.14 |
-16.35 |
2 |
10 |
0 |
137 |
328.277 |
6 |
↓
|
|
|
|
Analogs
-
11990270
-
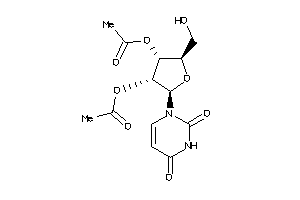
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.64 |
-3.37 |
-18.2 |
2 |
10 |
0 |
137 |
328.277 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
36144089
-
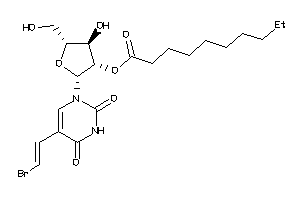
-
36144070
-
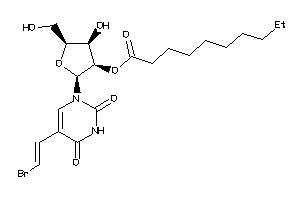
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KITM-1-E |
Thymidine Kinase, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.25 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KITM_HUMAN |
O00142
|
Thymidine Kinase, Mitochondrial, Human |
6300 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
3.62 |
-18.12 |
3 |
9 |
0 |
131 |
475.336 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.20 |
0.97 |
-53.59 |
2 |
9 |
-1 |
134 |
474.328 |
11 |
↓
|
|
|
|
Analogs
-
36144070
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KITM-1-E |
Thymidine Kinase, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6800 |
0.23 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KITM_HUMAN |
O00142
|
Thymidine Kinase, Mitochondrial, Human |
6800 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.76 |
5.19 |
-18.11 |
3 |
9 |
0 |
131 |
503.39 |
13 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.21 |
2.53 |
-53.54 |
2 |
9 |
-1 |
134 |
502.382 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
2400 |
0.28 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547_9HIV1 |
Q72547
|
Human Immunodeficiency Virus Type 1 Reverse Transcriptase, 9hiv1 |
10000 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.28 |
-3.69 |
-376.9 |
3 |
16 |
-4 |
258 |
463.125 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.28 |
-4.85 |
-244.57 |
4 |
16 |
-3 |
255 |
464.133 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
87 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.99 |
-5.46 |
-248.47 |
3 |
18 |
-3 |
285 |
489.147 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.99 |
-4.35 |
-387.9 |
2 |
18 |
-4 |
288 |
488.139 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
4 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.79 |
-2.39 |
-381.11 |
1 |
18 |
-4 |
285 |
489.123 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.79 |
-3.55 |
-241.1 |
2 |
18 |
-3 |
282 |
490.131 |
8 |
↓
|
|
|
|
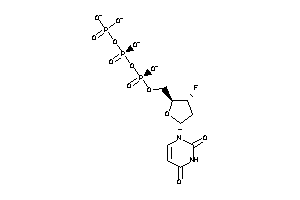
Analogs
-
36152111
-
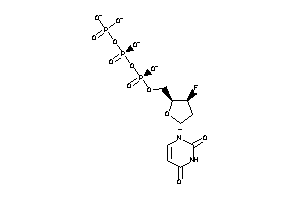
-
13517245
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPOG1-1-E |
DNA Polymerase Gamma Subunit 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.27 |
Binding ≤ 10μM
|
|
DPOLB-1-E |
DNA Polymerase Beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.28 |
Binding ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
70 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.88 |
-0.09 |
-356.8 |
1 |
15 |
-4 |
235 |
466.1 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.88 |
-1.25 |
-221.17 |
2 |
15 |
-3 |
232 |
467.108 |
8 |
↓
|
|
|
|
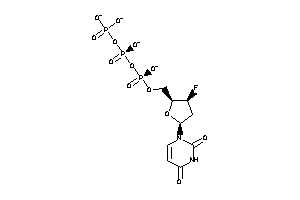
Analogs
-
36152106
-
-
13517245
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPOG1-1-E |
DNA Polymerase Gamma Subunit 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.27 |
Binding ≤ 10μM
|
|
DPOLB-1-E |
DNA Polymerase Beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.28 |
Binding ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
70 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.88 |
0.85 |
-368.88 |
1 |
15 |
-4 |
235 |
466.1 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.88 |
-0.31 |
-231.87 |
2 |
15 |
-3 |
232 |
467.108 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
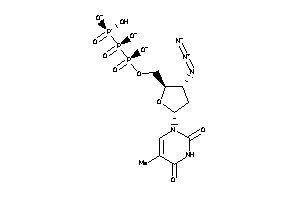
Analogs
-
36152231
-
-
36152211
-
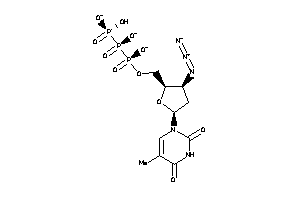
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
34 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.70 |
-6.5 |
-223.24 |
2 |
16 |
-3 |
264 |
472.16 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.70 |
-5.39 |
-380.93 |
1 |
16 |
-4 |
267 |
471.152 |
7 |
↓
|
|