|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
3830577
-
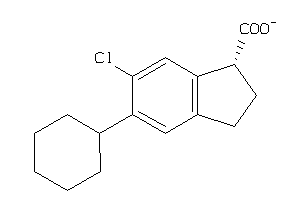
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
1 |
-50.26 |
0 |
2 |
-1 |
40 |
277.771 |
2 |
↓
|
|
|
|
Analogs
-
402782
-
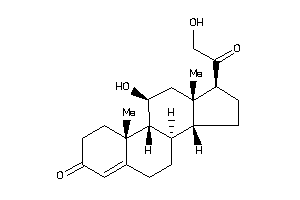
-
4095869
-
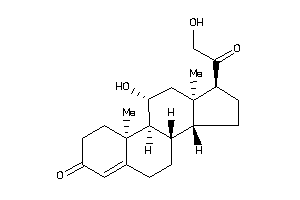
-
4095870
-
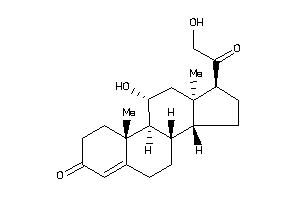
-
4095871
-
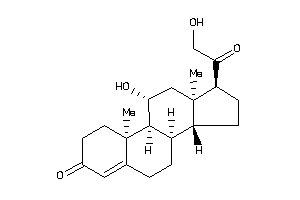
-
4095872
-
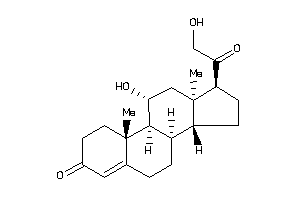
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9R,10S,11R,13S,14S,17R)-17-glycoloyl-11-hydroxy-10,13-dimethyl-1,2,6,7,8,9,11,12,14,15,16,17-dod
(8R,9R,10S,11R,13S,14S,17R)-17-g…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
-1.71 |
-10.96 |
2 |
4 |
0 |
74 |
346.467 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
401600
-
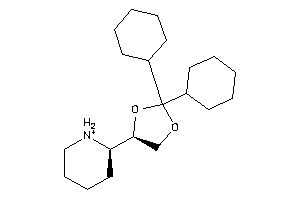
-
2036996
-
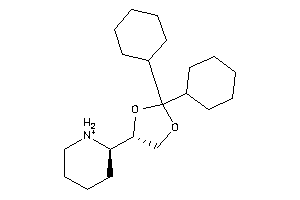
-
2036997
-
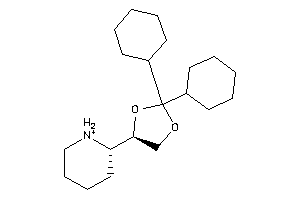
Draw
Identity
99%
90%
80%
70%
Popular Name:
1,3-Dioxolane, 2,2-dicyclohexyl-4-(2-piperidyl)-, hydrochloride, beta-; LS-62533; beta-2,2-Dicyclohexyl-4-(2-piperidyl)-1,3-dioxolane hydrochloride
1,3-Dioxolane, 2,2-dicyclohexyl-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.43 |
9.91 |
-42.27 |
2 |
3 |
1 |
35 |
322.513 |
3 |
↓
|
|
|
|
|
|
|
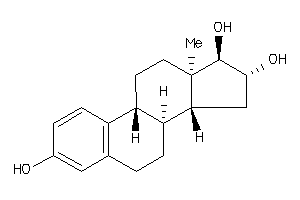
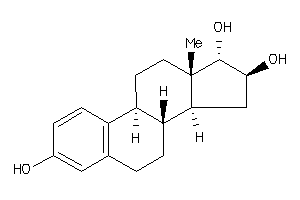
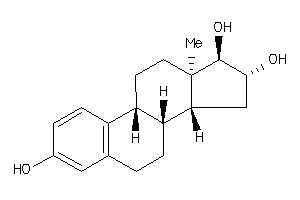
Analogs
-
1373
-
-
8866
-
-
525563
-
-
525565
-
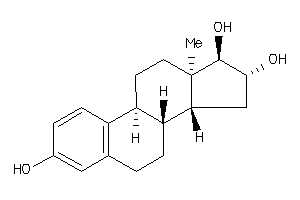
-
525566
-
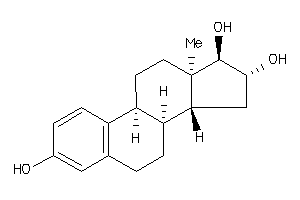
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,13R,14R,16S,17R)-13-methyl-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthrene-3,16,
(8S,9R,13R,14R,16S,17R)-13-methy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
-4.75 |
-6.17 |
3 |
3 |
0 |
60 |
288.387 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
1869701
-
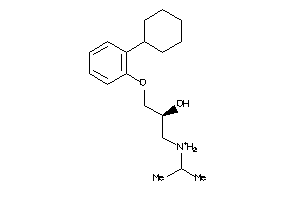
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-2.08 |
-39.72 |
3 |
3 |
1 |
46 |
292.443 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
2008383
-
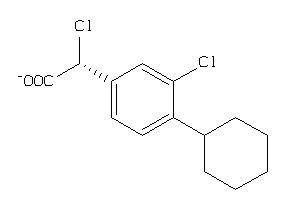
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
0.94 |
-43.9 |
0 |
2 |
-1 |
40 |
286.178 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
897263
-
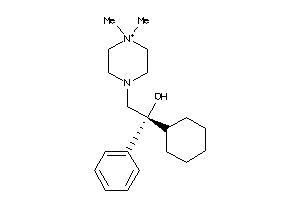
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.43 |
9.3 |
-31.6 |
1 |
3 |
1 |
23 |
317.497 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.43 |
10.45 |
-108.04 |
2 |
3 |
2 |
25 |
318.505 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
2034892
-
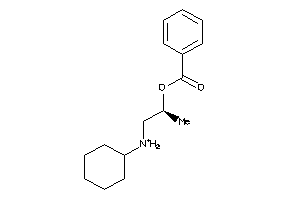
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN1A-2-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
|
SCN3A-2-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
|
SCN8A-2-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.16 |
9.32 |
-39.71 |
2 |
3 |
1 |
43 |
262.373 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
-0.23 |
-57.68 |
2 |
6 |
-1 |
102 |
325.41 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.13 |
1.71 |
-14.43 |
3 |
6 |
0 |
95 |
326.418 |
4 |
↓
|
|
|
|
Analogs
-
5759351
-
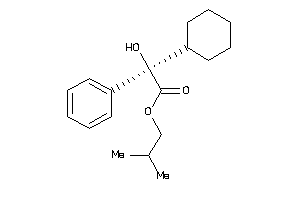
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.61 |
1.2 |
-4.89 |
1 |
3 |
0 |
46 |
290.403 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
3789310
-
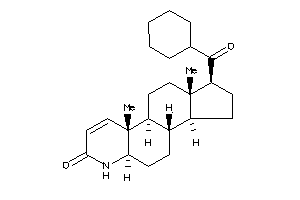
-
3789315
-
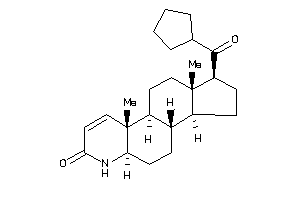
-
3791185
-
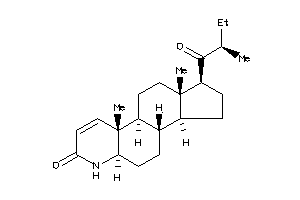
-
3791186
-
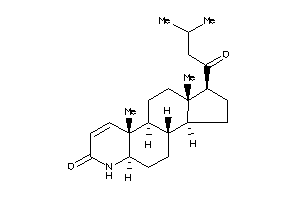
-
5440632
-
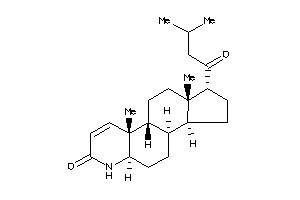
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bS,5aR,9aR,9bS,11aS)-1-butyryl-9a,11a-dimethyl-1,2,3,3a,3b,4,5,5a,6,9b,10,11-dodecahydroind
(1R,3aS,3bS,5aR,9aR,9bS,11aS)-1-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
0.4 |
-10.86 |
1 |
3 |
0 |
46 |
343.511 |
3 |
↓
|
|
|
|
Analogs
-
1846404
-
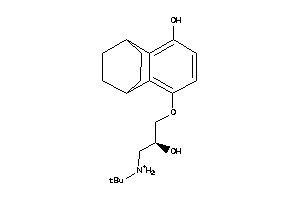
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
-4.14 |
-41.18 |
4 |
4 |
1 |
66 |
320.453 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
4214159
-
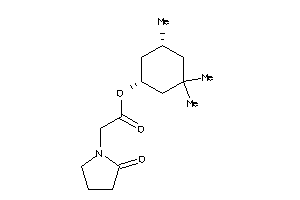
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
2.47 |
-11.7 |
0 |
4 |
0 |
46 |
267.369 |
4 |
↓
|
|
|
|
Analogs
-
403566
-
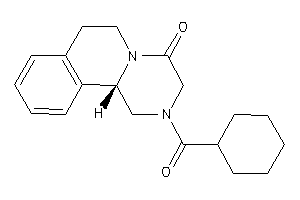
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
8.72 |
-10.26 |
0 |
4 |
0 |
41 |
312.413 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
1846186
-
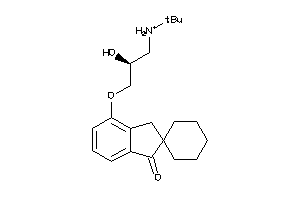
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
-1.85 |
-46.11 |
3 |
4 |
1 |
63 |
346.491 |
6 |
↓
|
|
|
|
Analogs
-
1842638
-
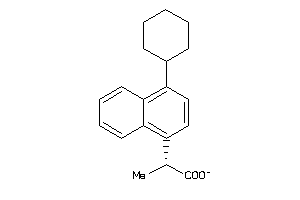
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.69 |
1.77 |
-51.07 |
0 |
2 |
-1 |
40 |
281.375 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
2200
-
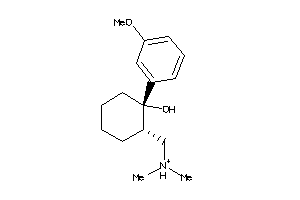
-
3051
-
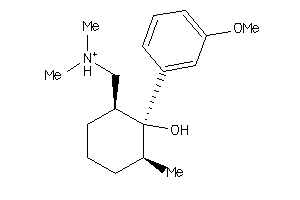
-
488880
-
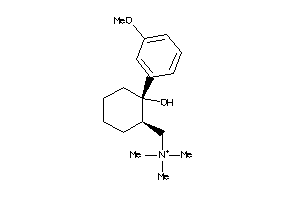
-
488881
-
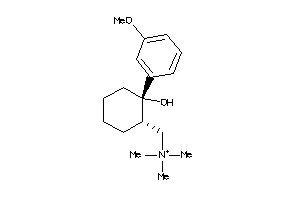
-
1849532
-
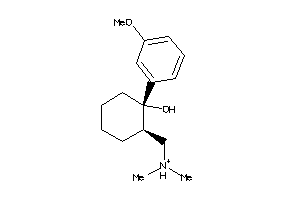
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.59 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
14 |
0.58 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5300 |
0.39 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7600 |
0.38 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
790 |
0.45 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.42 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.40 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.40 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1100 |
0.44 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1900 |
0.42 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3100 |
0.41 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4300 |
0.40 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu Opioid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.42 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
9.4 |
0.59 |
Binding ≤ 1μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
14 |
0.58 |
Binding ≤ 1μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
790 |
0.45 |
Binding ≤ 1μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
9.4 |
0.59 |
Binding ≤ 10μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
14 |
0.58 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
1600 |
0.43 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
2400 |
0.41 |
Binding ≤ 10μM |
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
3861 |
0.40 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
1493 |
0.43 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
2000 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
6.41 |
-36.54 |
2 |
3 |
1 |
34 |
264.389 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
-1.63 |
-12.71 |
2 |
5 |
0 |
61 |
423.532 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
-3.92 |
-14.56 |
2 |
6 |
0 |
70 |
399.535 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
-4.01 |
-13.77 |
2 |
6 |
0 |
70 |
399.535 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.60 |
9.29 |
-18.59 |
2 |
8 |
0 |
83 |
520.674 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.60 |
9.25 |
-18.34 |
2 |
8 |
0 |
83 |
520.674 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.71 |
-3.14 |
-16.43 |
1 |
5 |
0 |
52 |
383.536 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.46 |
10.84 |
-13.56 |
1 |
5 |
0 |
53 |
411.59 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
-4.2 |
-13.77 |
3 |
6 |
0 |
73 |
386.54 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
10.9 |
-16.95 |
3 |
6 |
0 |
73 |
448.611 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-4.27 |
-14.12 |
3 |
6 |
0 |
73 |
372.513 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
8.65 |
-18.9 |
3 |
6 |
0 |
73 |
400.567 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
-5.17 |
-14.68 |
2 |
6 |
0 |
64 |
398.551 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
-4.42 |
-12.4 |
2 |
6 |
0 |
64 |
426.605 |
4 |
↓
|
|