|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.18 |
4.66 |
-40.57 |
2 |
5 |
1 |
63 |
247.303 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.64 |
2.07 |
-49.81 |
0 |
5 |
-1 |
65 |
245.287 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.18 |
4.22 |
-15.9 |
1 |
5 |
0 |
62 |
246.295 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
6.79 |
-35.7 |
1 |
9 |
0 |
114 |
471.564 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.15 |
7.19 |
-54.82 |
2 |
9 |
1 |
116 |
472.572 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
52 |
0.32 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
52 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.07 |
14.2 |
-44.04 |
1 |
6 |
1 |
65 |
442.568 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.07 |
13.77 |
-16.29 |
0 |
6 |
0 |
64 |
441.56 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.07 |
14.56 |
-93.74 |
2 |
6 |
2 |
66 |
443.576 |
6 |
↓
|
|
|
|
Analogs
-
65361829
-
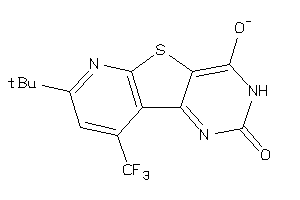
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
2.23 |
-39.64 |
1 |
5 |
-1 |
82 |
328.295 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.12 |
-2.8 |
-42.24 |
3 |
5 |
1 |
80 |
330.311 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.12 |
3.65 |
-7.28 |
2 |
5 |
0 |
79 |
329.303 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
9.22 |
-18.68 |
1 |
7 |
0 |
86 |
471.464 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.48 |
8.33 |
-48.65 |
0 |
7 |
-1 |
89 |
470.456 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
99 |
0.41 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
50 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
7.88 |
-66.62 |
1 |
6 |
0 |
75 |
338.392 |
2 |
↓
|
|
|
|
Analogs
-
34600443
-
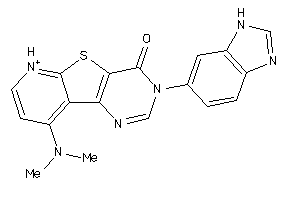
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
23 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
8.35 |
-41.46 |
1 |
5 |
1 |
52 |
341.391 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1250 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
8.91 |
-41.08 |
1 |
5 |
1 |
52 |
402.297 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
32 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
9.29 |
-42.13 |
1 |
5 |
1 |
52 |
391.398 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.47 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2440 |
0.31 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
9.96 |
-40.68 |
1 |
5 |
1 |
52 |
351.455 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.41 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2300 |
0.32 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
45 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.02 |
9.61 |
-40.74 |
1 |
5 |
1 |
52 |
392.291 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
39 |
0.41 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
39 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
4.98 |
-42.13 |
1 |
7 |
1 |
74 |
354.415 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.06 |
5.69 |
-83.32 |
2 |
7 |
2 |
76 |
355.423 |
3 |
↓
|
|
|
|
Analogs
-
34600443
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
62 |
0.39 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1180 |
0.32 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
122 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
6.93 |
-44.5 |
2 |
7 |
1 |
81 |
363.426 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
251 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
251 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
9.76 |
-40.04 |
1 |
5 |
1 |
52 |
351.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.45 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
613 |
0.35 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
9 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
9.73 |
-40.74 |
1 |
5 |
1 |
52 |
351.455 |
3 |
↓
|
|
|
|
Analogs
-
34600443
-

-
184798
-
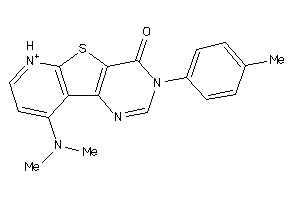
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.46 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
194 |
0.38 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
442 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
9.72 |
-40.51 |
1 |
5 |
1 |
52 |
351.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
180 |
0.36 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
180 |
0.36 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2040 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
10.5 |
-40.45 |
1 |
5 |
1 |
52 |
365.482 |
4 |
↓
|
|
|
|
Analogs
-
34600443
-

-
184798
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
407 |
0.34 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
407 |
0.34 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3540 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
10.2 |
-40.45 |
1 |
5 |
1 |
52 |
365.482 |
3 |
↓
|
|
|
|
Analogs
-
184798
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.26 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8500 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.23 |
10.6 |
-40.35 |
1 |
5 |
1 |
52 |
379.509 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
87 |
0.38 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
87 |
0.38 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4620 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
10.39 |
-40.72 |
1 |
5 |
1 |
52 |
363.466 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
12.08 |
-40.29 |
1 |
5 |
1 |
52 |
405.547 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
632 |
0.43 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
632 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.62 |
6.27 |
-40.76 |
1 |
5 |
1 |
52 |
287.368 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
167 |
0.45 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
167 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
6.6 |
-40.41 |
1 |
5 |
1 |
52 |
301.395 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1160 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
7.24 |
-39.68 |
1 |
5 |
1 |
52 |
315.422 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
650 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
9.47 |
-39.1 |
1 |
5 |
1 |
52 |
329.449 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
130 |
0.40 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
130 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
7.87 |
-38.77 |
1 |
5 |
1 |
52 |
343.476 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
130 |
0.40 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
130 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
8.92 |
-40.03 |
1 |
5 |
1 |
52 |
343.476 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
130 |
0.40 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
130 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
8.92 |
-39.99 |
1 |
5 |
1 |
52 |
343.476 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
130 |
0.40 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
130 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
7.87 |
-38.82 |
1 |
5 |
1 |
52 |
343.476 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
482 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
482 |
0.37 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2160 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
7.5 |
-38.71 |
1 |
5 |
1 |
52 |
343.476 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
482 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
482 |
0.37 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2160 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
8.57 |
-40.11 |
1 |
5 |
1 |
52 |
343.476 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
482 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
482 |
0.37 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2160 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
8.57 |
-40.14 |
1 |
5 |
1 |
52 |
343.476 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
482 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
482 |
0.37 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2160 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
7.49 |
-38.71 |
1 |
5 |
1 |
52 |
343.476 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
140 |
0.40 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
140 |
0.40 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
592 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
8.78 |
-42.01 |
1 |
5 |
1 |
52 |
343.476 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.93 |
8.34 |
-14.54 |
0 |
5 |
0 |
51 |
342.468 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
147 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
8.46 |
-40.12 |
1 |
5 |
1 |
52 |
343.476 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.47 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.47 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
342 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
6.77 |
-40.44 |
1 |
6 |
1 |
56 |
344.464 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
13 |
0.44 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
13 |
0.44 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
179 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
8.87 |
-40.29 |
1 |
5 |
1 |
52 |
357.503 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
76 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
76 |
0.37 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
154 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
9.62 |
-39.69 |
1 |
5 |
1 |
52 |
381.525 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
335 |
0.41 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
335 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
8.08 |
-40.4 |
1 |
5 |
1 |
52 |
317.438 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.46 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
26 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
8.43 |
-40.34 |
1 |
5 |
1 |
52 |
329.449 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
139 |
0.40 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
139 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
10.52 |
-39.64 |
1 |
5 |
1 |
52 |
343.476 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
44 |
0.40 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
44 |
0.40 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
344 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
9.61 |
-40.19 |
1 |
5 |
1 |
52 |
365.482 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
84 |
0.37 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4530 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
10.64 |
-39.64 |
1 |
5 |
1 |
52 |
379.509 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
101 |
0.34 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
101 |
0.34 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
901 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.16 |
12.16 |
-39.37 |
1 |
5 |
1 |
52 |
407.563 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.40 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
37 |
0.40 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2570 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
10.47 |
-40.44 |
1 |
5 |
1 |
52 |
365.482 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
71 |
0.36 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
71 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
9.56 |
-42.35 |
1 |
6 |
1 |
62 |
395.508 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
113 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
113 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
9.54 |
-39.89 |
1 |
5 |
1 |
52 |
363.466 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
41 |
0.40 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
41 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
10.9 |
-37.82 |
2 |
5 |
1 |
61 |
363.466 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.39 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
49 |
0.39 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2100 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
9.55 |
-20.22 |
1 |
6 |
0 |
84 |
361.43 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
99 |
0.35 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
99 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
10.48 |
-43.82 |
2 |
5 |
1 |
61 |
405.425 |
5 |
↓
|
|