|
|
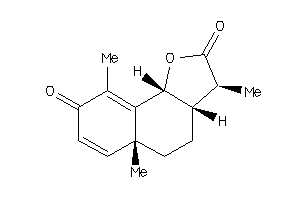
Analogs
-
119967
-
-
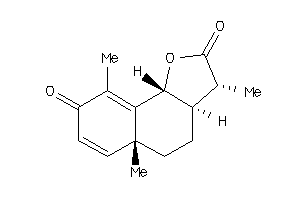
155356
-
-
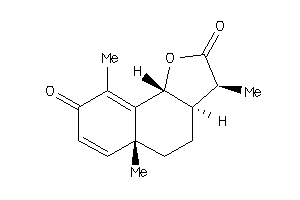
525571
-
-
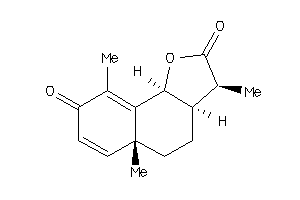
1069133
-
-
3881689
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
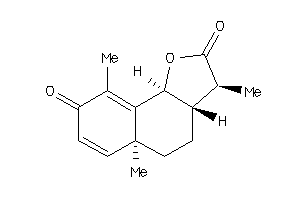
3,5a,9-trimethyl-3a,5,5a,9b-tetrahydronaphtho[1,2-b]furan-2,8(3H,4H)-dione
3,5a,9-trimethyl-3a,5,5a,9b-tetr…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
7.84 |
-13.76 |
0 |
3 |
0 |
43 |
246.306 |
0 |
↓
|
|
|
|
Analogs
-
31284427
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
9.37 |
-14.1 |
1 |
7 |
0 |
99 |
476.541 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8S,9S,10S,13S,14S)-13-methyl-3,17-dioxo-7,8,9,11,12,14,15,16-octahydro-6H-cyclopenta[a]phenanthren
[(8S,9S,10S,13S,14S)-13-methyl-3…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
9.65 |
-15.23 |
0 |
4 |
0 |
60 |
328.408 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
5.23 |
-7.9 |
1 |
2 |
0 |
37 |
232.323 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
6.88 |
-126.45 |
2 |
7 |
-2 |
138 |
380.352 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.31 |
11.65 |
-233.44 |
0 |
7 |
-3 |
137 |
379.344 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.06 |
9.59 |
-258.91 |
1 |
7 |
-3 |
141 |
379.344 |
5 |
↓
|
|
|
|
Analogs
-
5758815
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9S,10R,13S,14S)-10,13-dimethyl-17-methylene-7,8,9,11,12,14,15,16-octahydro-6H-cyclopenta[a]phena
(8S,9S,10R,13S,14S)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
10.57 |
-7.97 |
0 |
1 |
0 |
17 |
282.427 |
0 |
↓
|
|
|
|
Analogs
-
12496631
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6R,8S,9S,10R,13S,14S)-6-hydroxy-10,13-dimethyl-17-methylene-7,8,9,11,12,14,15,16-octahydro-6H-cyclo
(6R,8S,9S,10R,13S,14S)-6-hydroxy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
7.6 |
-6.98 |
1 |
2 |
0 |
37 |
298.426 |
0 |
↓
|
|
|
|
Analogs
-
12496631
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(6R,8S,9S,10R,13R,14S)-6-hydroxy-10,13-dimethyl-6,7,8,9,11,12,14,15-octahydrocyclopenta[a]phenanthre
(6R,8S,9S,10R,13R,14S)-6-hydroxy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
7.04 |
-7.04 |
1 |
2 |
0 |
37 |
284.399 |
0 |
↓
|
|
|
|
Analogs
-
968376
-
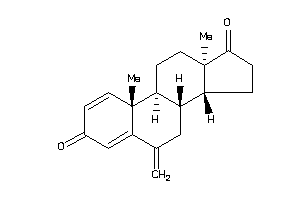
-
3830821
-
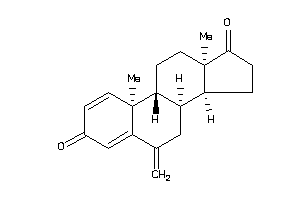
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9S,10R,13R,14S)-10,13-dimethyl-6-methylene-7,8,9,11,12,14,15,16-octahydrocyclopenta[a]phenanthre
(8S,9S,10R,13R,14S)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
9.5 |
-11.96 |
0 |
2 |
0 |
34 |
296.41 |
0 |
↓
|
|
|
|
Analogs
-
538220
-
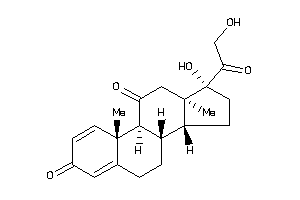
-
3831035
-
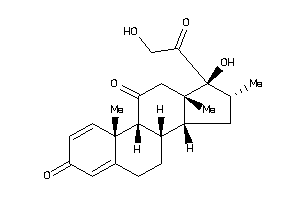
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9S,10S,13S,14R,17S)-17-hydroxy-17-(2-hydroxyacetyl)-10,13-dimethyl-6,7,8,9,12,14,15,16-octahydro
(8S,9S,10S,13S,14R,17S)-17-hydro…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
4.69 |
-17.22 |
2 |
5 |
0 |
92 |
358.434 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[2-[(8S,9S,10R,11S,13S,14S,16R,17R)-11,17-dihydroxy-10,13,16-trimethyl-3-oxo-7,8,9,11,12,14,15,16-oc
[2-[(8S,9S,10R,11S,13S,14S,16R,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
7.92 |
-14.81 |
2 |
6 |
0 |
101 |
416.514 |
4 |
↓
|
|
|
|
Analogs
-
8036054
-
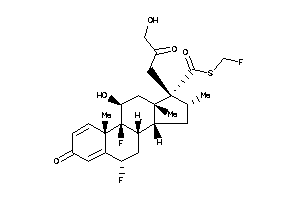
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
7.27 |
-21.05 |
2 |
5 |
0 |
92 |
500.579 |
6 |
↓
|
|
|
|
Analogs
-
8036054
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
7.38 |
-20.46 |
2 |
5 |
0 |
92 |
500.579 |
6 |
↓
|
|
|
|
Analogs
-
8036054
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
8.01 |
-22.19 |
2 |
5 |
0 |
92 |
500.579 |
6 |
↓
|
|
|
|
Analogs
-
8036054
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
7.25 |
-22.74 |
2 |
5 |
0 |
92 |
500.579 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
12.15 |
-17.01 |
0 |
6 |
0 |
86 |
484.976 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3aR,6Z,9S,9aR,10R,11aS)-6-(diethylaminomethylene)-1,5-dihydroxy-9-(methoxymethyl)-9a,11a-dimeth
[(1S,3aR,6Z,9S,9aR,10R,11aS)-6-(…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.23 |
7.73 |
-14.36 |
2 |
9 |
0 |
123 |
503.592 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.23 |
8.78 |
-53.01 |
1 |
9 |
-1 |
125 |
502.584 |
7 |
↓
|
|
|
|
Analogs
-
38269708
-
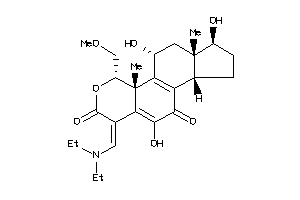
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,6Z,9S,9aR,10R,11aS)-6-(diethylaminomethylene)-1,5,10-trihydroxy-9-(methoxymethyl)-9a,11a-dim
(1S,3aR,6Z,9S,9aR,10R,11aS)-6-(d…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
4.12 |
-15.91 |
3 |
8 |
0 |
117 |
461.555 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.53 |
5.7 |
-108.36 |
1 |
8 |
-2 |
122 |
459.539 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.53 |
4.92 |
-62.01 |
2 |
8 |
-1 |
119 |
460.547 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3aS,6Z,9S,9aR,10R,11aS)-6-[[tert-butyl(methyl)amino]methylene]-1,5-dihydroxy-9-(methoxymethyl)-
[(1S,3aS,6Z,9S,9aR,10R,11aS)-6-[…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
8.7 |
-58.42 |
1 |
9 |
-1 |
125 |
516.611 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.67 |
7.88 |
-13.4 |
2 |
9 |
0 |
123 |
517.619 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3aR,6Z,9S,9aR,10R,11aS)-6-[[tert-butyl(methyl)amino]methylene]-1,5-dihydroxy-9-(methoxymethyl)-
[(1S,3aR,6Z,9S,9aR,10R,11aS)-6-[…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
8.68 |
-55.56 |
1 |
9 |
-1 |
125 |
516.611 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.67 |
7.85 |
-12.94 |
2 |
9 |
0 |
123 |
517.619 |
6 |
↓
|
|
|
|
Analogs
-
42875406
-
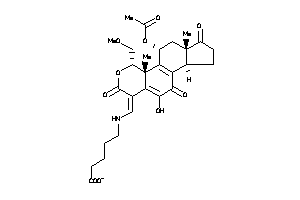
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3aS,6Z,9S,9aR,10R,11aS)-6-[[butyl(methyl)amino]methylene]-1,5-dihydroxy-9-(methoxymethyl)-9a,11
[(1S,3aS,6Z,9S,9aR,10R,11aS)-6-[…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
9.46 |
-54.46 |
1 |
9 |
-1 |
125 |
516.611 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.92 |
8.67 |
-11.37 |
2 |
9 |
0 |
123 |
517.619 |
8 |
↓
|
|
|
|
Analogs
-
42875406
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3aR,6Z,9S,9aR,10R,11aS)-6-[[butyl(methyl)amino]methylene]-1,5-dihydroxy-9-(methoxymethyl)-9a,11
[(1S,3aR,6Z,9S,9aR,10R,11aS)-6-[…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
9.41 |
-51.86 |
1 |
9 |
-1 |
125 |
516.611 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.92 |
8.6 |
-11.04 |
2 |
9 |
0 |
123 |
517.619 |
8 |
↓
|
|
|
|
Analogs
-
38270675
-
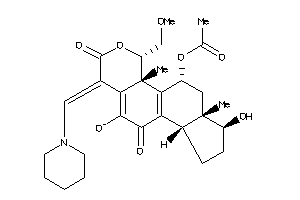
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3aS,6Z,9S,9aR,10R,11aS)-1,5-dihydroxy-9-(methoxymethyl)-9a,11a-dimethyl-4,7-dioxo-6-(pyrrolidin
[(1S,3aS,6Z,9S,9aR,10R,11aS)-1,5…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
7.25 |
-14.14 |
2 |
9 |
0 |
123 |
501.576 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.88 |
8.05 |
-58.99 |
1 |
9 |
-1 |
125 |
500.568 |
5 |
↓
|
|
|
|
Analogs
-
38270675
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3aR,6Z,9S,9aR,10R,11aS)-1,5-dihydroxy-9-(methoxymethyl)-9a,11a-dimethyl-4,7-dioxo-6-(pyrrolidin
[(1S,3aR,6Z,9S,9aR,10R,11aS)-1,5…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
7.23 |
-13.74 |
2 |
9 |
0 |
123 |
501.576 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.88 |
8 |
-56.16 |
1 |
9 |
-1 |
125 |
500.568 |
5 |
↓
|
|
|
|
Analogs
-
35904345
-
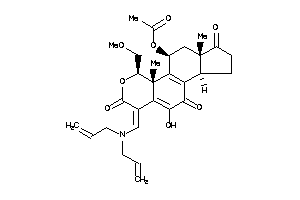
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3aS,6E,9S,9aR,10R,11aS)-6-[(diallylamino)methylene]-5-hydroxy-9-(methoxymethyl)-9a,11a-dimethyl-1,
[(3aS,6E,9S,9aR,10R,11aS)-6-[(di…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
12.76 |
-43.19 |
0 |
9 |
-1 |
122 |
524.59 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.58 |
12.01 |
-13.67 |
1 |
9 |
0 |
119 |
525.598 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,6Z,9S,9aR,10R,11aS)-6-[[3-(dimethylamino)propyl-methyl-amino]methylene]-1,5,10-trihydroxy-9-
(1S,3aS,6Z,9S,9aR,10R,11aS)-6-[[…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.08 |
6.89 |
-76.26 |
3 |
9 |
0 |
124 |
504.624 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.08 |
6.09 |
-52.31 |
4 |
9 |
1 |
121 |
505.632 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.08 |
6.04 |
-71.93 |
3 |
9 |
0 |
124 |
504.624 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3aS,6E,9S,9aR,10R,11aS)-1,5-dihydroxy-9-(methoxymethyl)-9a,11a-dimethyl-6-(morpholinomethylene)
[(1S,3aS,6E,9S,9aR,10R,11aS)-1,5…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.33 |
7.14 |
-54.23 |
1 |
10 |
-1 |
135 |
516.567 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.33 |
6.32 |
-14.7 |
2 |
10 |
0 |
132 |
517.575 |
5 |
↓
|
|
|
|
Analogs
-
36159590
-
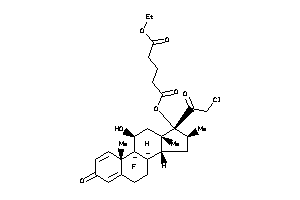
-
36159593
-
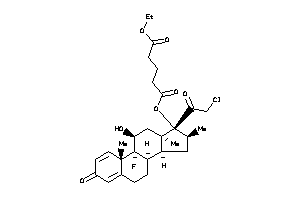
-
36159597
-
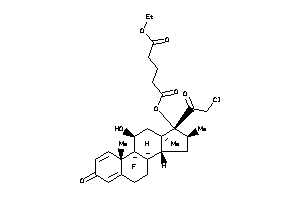
-
38432300
-
-
43279037
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
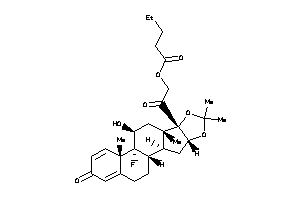
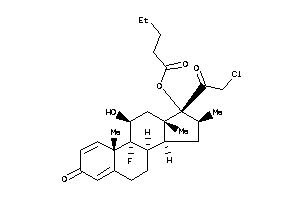
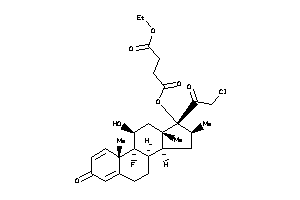
O5-[(8R,9R,10S,11S,13S,14S,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O5-[(8R,9R,10S,11S,13S,14S,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
12.98 |
-22.22 |
1 |
7 |
0 |
107 |
553.067 |
10 |
↓
|
|
|
|
Analogs
-
36159593
-

-
36159597
-

-
38432300
-

-
43279037
-

-
43279039
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
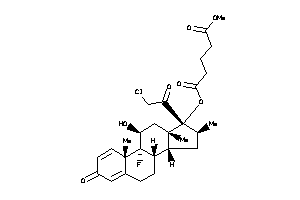
O5-[(8R,9R,10S,11S,13S,14R,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O5-[(8R,9R,10S,11S,13S,14R,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
13.4 |
-21.16 |
1 |
7 |
0 |
107 |
553.067 |
10 |
↓
|
|
|
|
Analogs
-
36159597
-

-
38432300
-

-
43279037
-

-
43279039
-

-
43279041
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
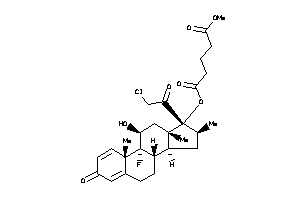
O5-[(8R,9R,10S,11S,13R,14S,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O5-[(8R,9R,10S,11S,13R,14S,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
13.57 |
-19.8 |
1 |
7 |
0 |
107 |
553.067 |
10 |
↓
|
|
|
|
Analogs
-
38432300
-

-
43279037
-

-
43279039
-

-
43279041
-

-
43279043
-
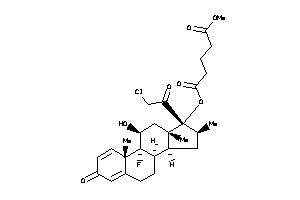
Draw
Identity
99%
90%
80%
70%
Popular Name:
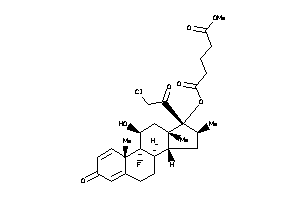
O5-[(8R,9R,10S,11S,13R,14R,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O5-[(8R,9R,10S,11S,13R,14R,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
12.3 |
-22.59 |
1 |
7 |
0 |
107 |
553.067 |
10 |
↓
|
|
|
|
Analogs
-
36159607
-
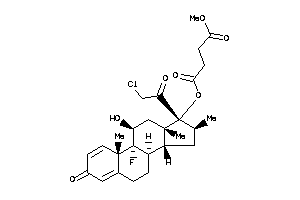
-
36159612
-
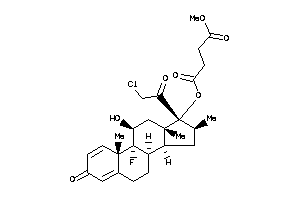
-
36159616
-
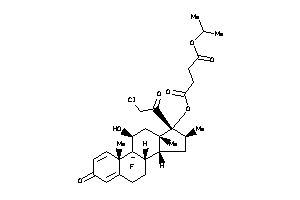
-
36159621
-
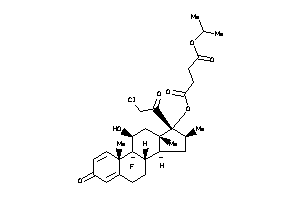
-
36159625
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(8S,9R,10S,11S,13S,14R,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O1-[(8S,9R,10S,11S,13S,14R,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
11.41 |
-17.47 |
1 |
7 |
0 |
107 |
525.013 |
8 |
↓
|
|
|
|
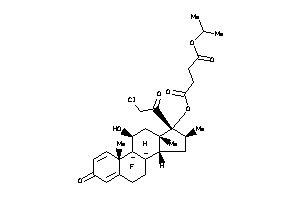
Analogs
-
36159612
-

-
36159616
-

-
36159621
-

-
36159625
-

-
36159629
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(8R,9R,10S,11S,13S,14R,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O1-[(8R,9R,10S,11S,13S,14R,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
11.72 |
-15.26 |
1 |
7 |
0 |
107 |
525.013 |
8 |
↓
|
|
|
|
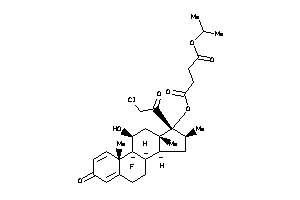
Analogs
-
36159616
-

-
36159621
-

-
36159625
-

-
36159629
-

-
43278823
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(8R,9R,10S,11S,13S,14S,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O1-[(8R,9R,10S,11S,13S,14S,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
11.31 |
-14.82 |
1 |
7 |
0 |
107 |
525.013 |
8 |
↓
|
|
|
|
Analogs
-
3924561
-
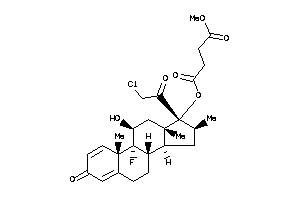
Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(8S,9R,10S,11S,13S,14R,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O1-[(8S,9R,10S,11S,13S,14R,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
13.04 |
-17.12 |
1 |
7 |
0 |
107 |
553.067 |
9 |
↓
|
|
|
|
Analogs
-
3924561
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(8S,9R,10S,11S,13S,14S,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O1-[(8S,9R,10S,11S,13S,14S,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
13.24 |
-16.24 |
1 |
7 |
0 |
107 |
553.067 |
9 |
↓
|
|
|
|
Analogs
-
3924561
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(8R,9R,10S,11S,13S,14R,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O1-[(8R,9R,10S,11S,13S,14R,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
13.35 |
-14.94 |
1 |
7 |
0 |
107 |
553.067 |
9 |
↓
|
|
|
|
Analogs
-
3924561
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(8R,9R,10S,11S,13S,14S,16S,17R)-17-(2-chloroacetyl)-9-fluoro-11-hydroxy-10,13,16-trimethyl-3-oxo
O1-[(8R,9R,10S,11S,13S,14S,16S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
12.94 |
-14.46 |
1 |
7 |
0 |
107 |
553.067 |
9 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-3-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.27 |
Binding ≤ 10μM
|
|
FNTB-3-E |
Protein Farnesyltransferase Beta Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
7.6 |
-23.54 |
0 |
7 |
0 |
88 |
428.481 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-3-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.27 |
Binding ≤ 10μM
|
|
FNTB-3-E |
Protein Farnesyltransferase Beta Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
7.73 |
-24.81 |
0 |
7 |
0 |
88 |
428.481 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-3-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.27 |
Binding ≤ 10μM
|
|
FNTB-3-E |
Protein Farnesyltransferase Beta Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
6.43 |
-20.59 |
0 |
7 |
0 |
88 |
428.481 |
1 |
↓
|
|
|
|
Analogs
-
1322099
-
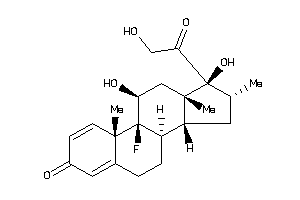
-
3830285
-
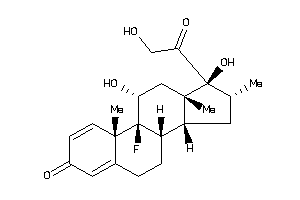
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
3.19 |
-12.39 |
3 |
5 |
0 |
95 |
392.467 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.99 |
2.5 |
-14.63 |
2 |
6 |
0 |
93 |
318.325 |
6 |
↓
|
|