|
|
Analogs
-
26257210
-
-
26264583
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
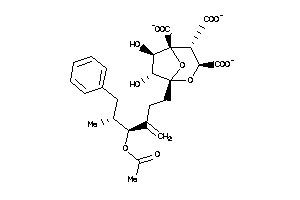
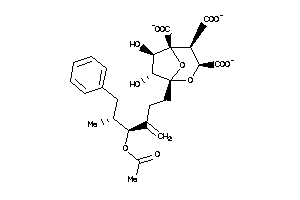
(1S,3R,4S,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,6,7-trihydroxy-2,8-d
(1S,3R,4S,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.30 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.30 |
7.5 |
-244.62 |
3 |
13 |
-3 |
226 |
535.478 |
12 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.30 |
5.59 |
-137.8 |
4 |
13 |
-2 |
223 |
536.486 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
57 |
0.28 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
11.54 |
-233.05 |
1 |
11 |
-3 |
185 |
503.48 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
57 |
0.28 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
13.36 |
-243.15 |
1 |
11 |
-3 |
185 |
503.48 |
12 |
↓
|
|
|
|
Analogs
-
43366097
-
-
43366099
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
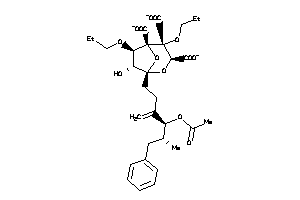
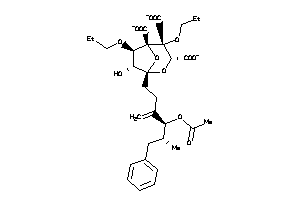
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-6-butoxy-4,7-dihydroxy
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
12.68 |
-259.61 |
2 |
13 |
-3 |
215 |
591.586 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.75 |
10.55 |
-130.74 |
3 |
13 |
-2 |
212 |
592.594 |
16 |
↓
|
|
|
|
Analogs
-
43366097
-

-
43366099
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-6-butoxy-4,7-dihydroxy
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
11.71 |
-257.79 |
2 |
13 |
-3 |
215 |
591.586 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.75 |
10.36 |
-129.41 |
3 |
13 |
-2 |
212 |
592.594 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-6-ethoxycarbonyloxy-4,
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
12.55 |
-253.58 |
2 |
15 |
-3 |
241 |
607.541 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.54 |
10.42 |
-127.28 |
3 |
15 |
-2 |
238 |
608.549 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-6-ethoxycarbonyloxy-4,
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
11.57 |
-252.03 |
2 |
15 |
-3 |
241 |
607.541 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.54 |
10.22 |
-126.15 |
3 |
15 |
-2 |
238 |
608.549 |
16 |
↓
|
|
|
|
Analogs
-
43366114
-
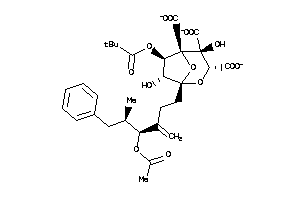
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-pentan
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
14.21 |
-255.46 |
2 |
14 |
-3 |
232 |
619.596 |
17 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.83 |
12.08 |
-127.63 |
3 |
14 |
-2 |
229 |
620.604 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-pentan
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
13.23 |
-253.8 |
2 |
14 |
-3 |
232 |
619.596 |
17 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.83 |
11.88 |
-126.42 |
3 |
14 |
-2 |
229 |
620.604 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4S,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-methox
(1S,3R,4S,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.17 |
11.23 |
-247.82 |
2 |
15 |
-3 |
241 |
593.514 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,4R,5R,6R,7R)-6-butanoyloxy-4,7-dihydroxy-1-[(4S,5R)-5-methyl-3-methylene-6-phenyl-4-propanoyl
(1S,3S,4R,5R,6R,7R)-6-butanoylox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
14.21 |
-255.45 |
2 |
14 |
-3 |
232 |
619.596 |
17 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.68 |
12.07 |
-127.44 |
3 |
14 |
-2 |
229 |
620.604 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4R,5R,6R,7R)-6-butanoyloxy-4,7-dihydroxy-1-[(4S,5R)-5-methyl-3-methylene-6-phenyl-4-propanoyl
(1S,3R,4R,5R,6R,7R)-6-butanoylox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
13.22 |
-253.76 |
2 |
14 |
-3 |
232 |
619.596 |
17 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.68 |
11.87 |
-126.32 |
3 |
14 |
-2 |
229 |
620.604 |
17 |
↓
|
|
|
|
Analogs
-
43366097
-

-
43366099
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-propox
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
11.9 |
-259.27 |
2 |
13 |
-3 |
215 |
577.559 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.20 |
9.76 |
-130.56 |
3 |
13 |
-2 |
212 |
578.567 |
15 |
↓
|
|
|
|
Analogs
-
43366097
-

-
43366099
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-propox
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
10.93 |
-257.3 |
2 |
13 |
-3 |
215 |
577.559 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.20 |
9.58 |
-129.17 |
3 |
13 |
-2 |
212 |
578.567 |
15 |
↓
|
|
|
|
Analogs
-
43366097
-

-
43366099
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4S,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-methox
(1S,3R,4S,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.32 |
9.86 |
-244.43 |
2 |
13 |
-3 |
215 |
549.505 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,4R,5R,6R,7R)-6-[(E,4S,6S)-4,6-dimethyloct-2-enoyl]oxy-4,7-dihydroxy-1-[(5S)-5-methyl-3-methyl
(1S,3S,4R,5R,6R,7R)-6-[(E,4S,6S)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
16.44 |
-256.64 |
2 |
12 |
-3 |
206 |
629.679 |
17 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.08 |
14.3 |
-126.87 |
3 |
12 |
-2 |
203 |
630.687 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4R,5R,6R,7R)-6-[(E,4S,6S)-4,6-dimethyloct-2-enoyl]oxy-4,7-dihydroxy-1-[(5S)-5-methyl-3-methyl
(1S,3R,4R,5R,6R,7R)-6-[(E,4S,6S)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
15.46 |
-254.53 |
2 |
12 |
-3 |
206 |
629.679 |
17 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.08 |
14.1 |
-125.54 |
3 |
12 |
-2 |
203 |
630.687 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-6-(ethylcarbamoyloxy)-
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.34 |
9.63 |
-256.09 |
3 |
15 |
-3 |
244 |
606.557 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.34 |
7.49 |
-128.8 |
4 |
15 |
-2 |
241 |
607.565 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-6-(ethylcarbamoyloxy)-
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.34 |
8.65 |
-254.43 |
3 |
15 |
-3 |
244 |
606.557 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.34 |
7.3 |
-127.74 |
4 |
15 |
-2 |
241 |
607.565 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4S,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-(3-met
(1S,3R,4S,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
13.5 |
-252.58 |
2 |
14 |
-3 |
232 |
617.58 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-[(2S)-
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
18 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
13.96 |
-257.91 |
2 |
14 |
-3 |
232 |
619.596 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.82 |
11.83 |
-129.11 |
3 |
14 |
-2 |
229 |
620.604 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-[(2S)-
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
18 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
12.98 |
-256.43 |
2 |
14 |
-3 |
232 |
619.596 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.82 |
11.62 |
-127.9 |
3 |
14 |
-2 |
229 |
620.604 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-propan
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
11 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
12.67 |
-255.52 |
2 |
14 |
-3 |
232 |
591.542 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.77 |
10.54 |
-127.81 |
3 |
14 |
-2 |
229 |
592.55 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydroxy-6-propan
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
11 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
11.69 |
-253.79 |
2 |
14 |
-3 |
232 |
591.542 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.77 |
10.33 |
-126.7 |
3 |
14 |
-2 |
229 |
592.55 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4S,5R,6R,7R)-6-acetoxy-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-4,7-dihydrox
(1S,3R,4S,5R,6R,7R)-6-acetoxy-1-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
36 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.40 |
11.53 |
-250.51 |
2 |
14 |
-3 |
232 |
577.515 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-6-butanoyloxy-4,7-dihy
(1S,3S,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
13.43 |
-255.45 |
2 |
14 |
-3 |
232 |
605.569 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.32 |
11.3 |
-127.59 |
3 |
14 |
-2 |
229 |
606.577 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4-acetoxy-5-methyl-3-methylene-6-phenyl-hexyl]-6-butanoyloxy-4,7-dihy
(1S,3R,4R,5R,6R,7R)-1-[(4S,5R)-4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
12.45 |
-253.76 |
2 |
14 |
-3 |
232 |
605.569 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.32 |
11.1 |
-126.44 |
3 |
14 |
-2 |
229 |
606.577 |
16 |
↓
|
|