|
|
Analogs
-
40874955
-
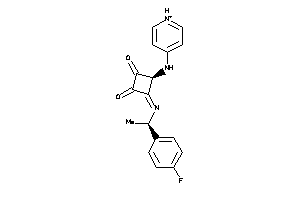
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8200 |
0.31 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
8200 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
5.38 |
-57.92 |
2 |
5 |
0 |
73 |
312.324 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.42 |
4.91 |
-8.87 |
1 |
5 |
0 |
71 |
311.316 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.42 |
5.33 |
-63.45 |
2 |
5 |
0 |
73 |
312.324 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.30 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.37 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.36 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.31 |
Binding ≤ 10μM
|
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.30 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5100 |
0.31 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8600 |
0.30 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
1.44 |
-76.04 |
4 |
7 |
0 |
116 |
323.332 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.39 |
0.96 |
-16.15 |
3 |
7 |
0 |
115 |
322.324 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.39 |
1.49 |
-61.13 |
4 |
7 |
0 |
116 |
323.332 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.30 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.37 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.36 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.31 |
Binding ≤ 10μM
|
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.30 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5100 |
0.31 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8600 |
0.30 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
1.49 |
-63.09 |
4 |
7 |
0 |
116 |
323.332 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.39 |
1.02 |
-13.16 |
3 |
7 |
0 |
115 |
322.324 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.39 |
1.44 |
-74.32 |
4 |
7 |
0 |
116 |
323.332 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.35 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
250 |
0.42 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
810 |
0.39 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8900 |
0.32 |
Binding ≤ 10μM
|
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.34 |
Binding ≤ 10μM
|
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.46 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
600 |
0.40 |
ADME/T ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3300 |
0.35 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1300 |
0.37 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KC1G1_HUMAN |
Q9HCP0
|
Casein Kinase I Gamma 1, Human |
250 |
0.42 |
Binding ≤ 1μM
|
|
ROCK1_HUMAN |
Q13464
|
Rho-associated Protein Kinase 1, Human |
60 |
0.46 |
Binding ≤ 1μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
810 |
0.39 |
Binding ≤ 1μM
|
|
KC1G1_HUMAN |
Q9HCP0
|
Casein Kinase I Gamma 1, Human |
250 |
0.42 |
Binding ≤ 10μM
|
|
MK01_HUMAN |
P28482
|
MAP Kinase ERK2, Human |
4600 |
0.34 |
Binding ≤ 10μM
|
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
8900 |
0.32 |
Binding ≤ 10μM
|
|
ROCK1_HUMAN |
Q13464
|
Rho-associated Protein Kinase 1, Human |
60 |
0.46 |
Binding ≤ 10μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
810 |
0.39 |
Binding ≤ 10μM
|
|
AURKB_HUMAN |
Q96GD4
|
Serine/threonine-protein Kinase Aurora-B, Human |
3200 |
0.35 |
Binding ≤ 10μM
|
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
600 |
0.40 |
ADME/T ≤ 10μM
|
|
CP2D6_HUMAN |
P10635
|
Cytochrome P450 2D6, Human |
3300 |
0.35 |
ADME/T ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
1300 |
0.37 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
5.31 |
-61.67 |
2 |
5 |
0 |
73 |
294.334 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.25 |
4.84 |
-8.35 |
1 |
5 |
0 |
71 |
293.326 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.25 |
5.25 |
-63.69 |
2 |
5 |
0 |
73 |
294.334 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
2.4 |
-63.46 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.75 |
1.93 |
-10.58 |
2 |
6 |
0 |
92 |
309.325 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.75 |
2.45 |
-62.46 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
2.44 |
-63.8 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.75 |
1.98 |
-10.46 |
2 |
6 |
0 |
92 |
309.325 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.75 |
2.39 |
-62.19 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
|
|
Analogs
-
40880512
-
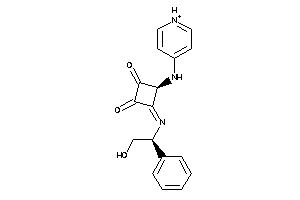
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5300 |
0.32 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
5300 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.25 |
2.13 |
-61.41 |
3 |
6 |
0 |
93 |
310.333 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.25 |
1.66 |
-10.07 |
2 |
6 |
0 |
92 |
309.325 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.25 |
2.07 |
-68.66 |
3 |
6 |
0 |
93 |
310.333 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.83 |
6.64 |
-63 |
1 |
5 |
0 |
64 |
308.361 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.83 |
6.17 |
-7.83 |
0 |
5 |
0 |
63 |
307.353 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.83 |
6.09 |
-8.46 |
0 |
5 |
0 |
63 |
307.353 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.29 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
3800 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
5.17 |
-16.76 |
2 |
7 |
0 |
101 |
350.378 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.82 |
5.11 |
-17.08 |
2 |
7 |
0 |
101 |
350.378 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.82 |
4.98 |
-66.73 |
3 |
7 |
0 |
102 |
351.386 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
5.97 |
-62.07 |
2 |
5 |
0 |
73 |
308.361 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.68 |
5.45 |
-9.02 |
1 |
5 |
0 |
71 |
307.353 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.68 |
5.92 |
-63.88 |
2 |
5 |
0 |
73 |
308.361 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
5.97 |
-61.63 |
2 |
5 |
0 |
73 |
308.361 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.68 |
5.44 |
-9.14 |
1 |
5 |
0 |
71 |
307.353 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.68 |
5.92 |
-64.49 |
2 |
5 |
0 |
73 |
308.361 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
3.18 |
-63.06 |
4 |
6 |
0 |
99 |
309.349 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.31 |
2.64 |
-9.68 |
3 |
6 |
0 |
97 |
308.341 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.31 |
3.12 |
-65.14 |
4 |
6 |
0 |
99 |
309.349 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.34 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
2300 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
5.56 |
-57.27 |
2 |
5 |
0 |
73 |
308.361 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.70 |
5.1 |
-7.99 |
1 |
5 |
0 |
71 |
307.353 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.70 |
5.54 |
-65.39 |
2 |
5 |
0 |
73 |
308.361 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.33 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.38 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.37 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
390 |
0.36 |
Binding ≤ 10μM
|
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4200 |
0.30 |
Binding ≤ 10μM
|
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.40 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6500 |
0.29 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KC1G1_HUMAN |
Q9HCP0
|
Casein Kinase I Gamma 1, Human |
160 |
0.38 |
Binding ≤ 1μM
|
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
390 |
0.36 |
Binding ≤ 1μM
|
|
ROCK1_HUMAN |
Q13464
|
Rho-associated Protein Kinase 1, Human |
60 |
0.40 |
Binding ≤ 1μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
290 |
0.37 |
Binding ≤ 1μM
|
|
KC1G1_HUMAN |
Q9HCP0
|
Casein Kinase I Gamma 1, Human |
160 |
0.38 |
Binding ≤ 10μM
|
|
MK01_HUMAN |
P28482
|
MAP Kinase ERK2, Human |
4200 |
0.30 |
Binding ≤ 10μM
|
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
390 |
0.36 |
Binding ≤ 10μM
|
|
ROCK1_HUMAN |
Q13464
|
Rho-associated Protein Kinase 1, Human |
60 |
0.40 |
Binding ≤ 10μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
290 |
0.37 |
Binding ≤ 10μM
|
|
AURKB_HUMAN |
Q96GD4
|
Serine/threonine-protein Kinase Aurora-B, Human |
1400 |
0.33 |
Binding ≤ 10μM
|
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
6500 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.89 |
-1.36 |
-60.98 |
5 |
8 |
0 |
136 |
339.331 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.89 |
-1.83 |
-14.11 |
4 |
8 |
0 |
135 |
338.323 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.89 |
-1.42 |
-74.77 |
5 |
8 |
0 |
136 |
339.331 |
5 |
↓
|
|
|
|
Analogs
-
40877622
-
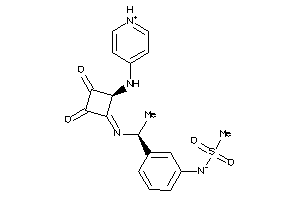
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.46 |
2.07 |
-69.14 |
2 |
8 |
0 |
121 |
386.433 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.46 |
1.54 |
-20.71 |
1 |
8 |
0 |
120 |
385.425 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.46 |
2.02 |
-62.03 |
2 |
8 |
0 |
121 |
386.433 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
5.32 |
-61.21 |
2 |
5 |
0 |
73 |
294.334 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.25 |
4.79 |
-9.25 |
1 |
5 |
0 |
71 |
293.326 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.25 |
5.26 |
-64.3 |
2 |
5 |
0 |
73 |
294.334 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.32 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
5000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.78 |
2.46 |
-61.13 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.78 |
1.99 |
-9.98 |
2 |
6 |
0 |
92 |
309.325 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.78 |
2.41 |
-64.87 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.32 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
5000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.78 |
2.45 |
-61.86 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.78 |
1.98 |
-10.1 |
2 |
6 |
0 |
92 |
309.325 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.78 |
2.4 |
-64.45 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
2.71 |
-65.43 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.20 |
2.18 |
-9.68 |
2 |
6 |
0 |
92 |
309.325 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.20 |
2.66 |
-63.27 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
2.64 |
-61.52 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.20 |
2.11 |
-9.8 |
2 |
6 |
0 |
92 |
309.325 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.20 |
2.59 |
-64.8 |
3 |
6 |
0 |
93 |
310.333 |
4 |
↓
|
|