|
|
|
|
|
Analogs
-
36488997
-
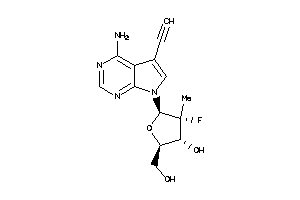
-
36152339
-
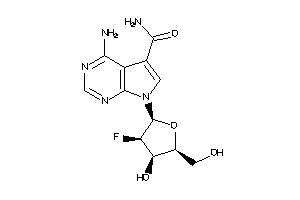
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.21 |
-4.34 |
-15.35 |
6 |
9 |
0 |
150 |
311.273 |
3 |
↓
|
|
|
|
Analogs
-
36152355
-
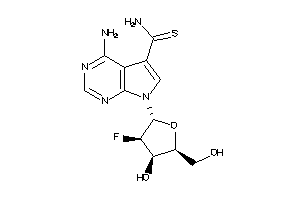
-
27416883
-
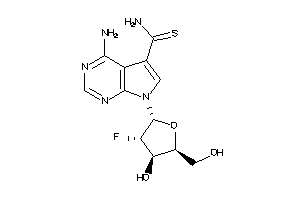
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.23 |
-1.88 |
-18.48 |
6 |
8 |
0 |
132 |
327.341 |
3 |
↓
|
|
|
|
Analogs
-
36152349
-
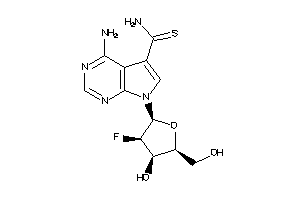
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.23 |
-2.09 |
-16.47 |
6 |
8 |
0 |
132 |
327.341 |
3 |
↓
|
|
|
|
Analogs
-
5162989
-
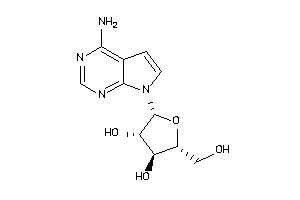
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.93 |
-3.53 |
-13.68 |
5 |
8 |
0 |
127 |
266.257 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.93 |
-3.27 |
-34.72 |
6 |
8 |
1 |
128 |
267.265 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.93 |
-6.99 |
-31.97 |
6 |
8 |
1 |
128 |
267.265 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADK-1-E |
Adenosine Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.23 |
-0.09 |
-9.5 |
4 |
7 |
0 |
106 |
362.127 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.23 |
0.18 |
-29.16 |
5 |
7 |
1 |
108 |
363.135 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.23 |
-4.21 |
-30.69 |
5 |
7 |
1 |
108 |
363.135 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.64 |
0.21 |
-141.55 |
3 |
10 |
-2 |
159 |
328.221 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.64 |
-0.94 |
-49.97 |
4 |
10 |
-1 |
156 |
329.229 |
4 |
↓
|
|
|
|
Analogs
-
38602735
-
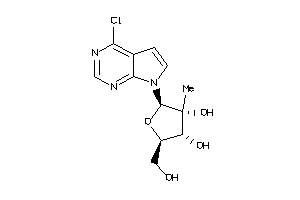
-
22054386
-
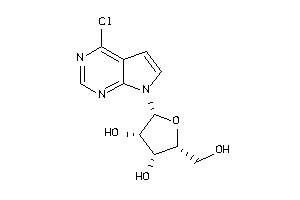
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.27 |
-2.28 |
-13.84 |
3 |
7 |
0 |
101 |
285.687 |
2 |
↓
|
|