|
|
Analogs
-
7997302
-
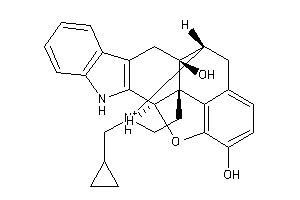
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.80 |
7.23 |
-55.86 |
4 |
5 |
1 |
70 |
415.513 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.80 |
4.98 |
-9.61 |
3 |
5 |
0 |
69 |
414.505 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.37 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
43 |
0.31 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
6.76 |
-52.69 |
4 |
6 |
1 |
79 |
445.539 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.81 |
5.21 |
-12.92 |
3 |
6 |
0 |
78 |
444.531 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.32 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
82 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
6.48 |
-89.46 |
9 |
8 |
2 |
134 |
473.577 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.71 |
4.96 |
-39.31 |
8 |
8 |
1 |
132 |
472.569 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
5.95 |
-53.19 |
8 |
8 |
1 |
134 |
472.569 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.35 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.32 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3911 |
0.24 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3911 |
0.24 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
411 |
0.28 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2510 |
0.25 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
40 |
0.32 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
6.57 |
0.36 |
Binding ≤ 1μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
550 |
0.27 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
411 |
0.28 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
6.57 |
0.36 |
Binding ≤ 10μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
40 |
0.32 |
Binding ≤ 10μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
1760 |
0.25 |
Binding ≤ 10μM
|
|
OPRK_CAVPO |
P41144
|
Kappa Opioid Receptor, Guinea Pig |
3911 |
0.24 |
Binding ≤ 10μM
|
|
OPRM_CAVPO |
P97266
|
Mu Opioid Receptor, Guinea Pig |
2510 |
0.25 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
550 |
0.27 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
2510 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
9.6 |
-50.86 |
3 |
5 |
1 |
59 |
429.54 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.11 |
8.01 |
-11.68 |
2 |
5 |
0 |
58 |
428.532 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
8.81 |
-56.92 |
3 |
6 |
1 |
76 |
471.577 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.96 |
6.66 |
-13.61 |
2 |
6 |
0 |
75 |
470.569 |
5 |
↓
|
|
|
|
Analogs
-
40918341
-
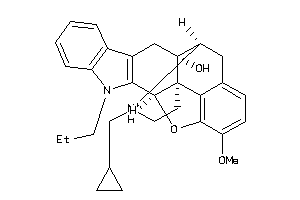
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.61 |
11.83 |
-57.39 |
3 |
6 |
1 |
83 |
510.658 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.61 |
9.68 |
-16.18 |
2 |
6 |
0 |
82 |
509.65 |
7 |
↓
|
|
|
|
Analogs
-
36221079
-
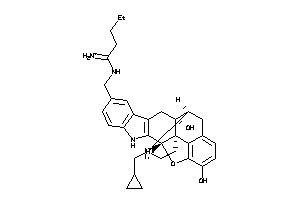
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
8.11 |
-99.46 |
7 |
7 |
2 |
108 |
514.67 |
7 |
↓
|
|
|
|
Analogs
-
36221079
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
7.39 |
-97.86 |
7 |
7 |
2 |
108 |
514.67 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
8.59 |
-53.77 |
3 |
5 |
1 |
59 |
429.54 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
9.27 |
-56.75 |
3 |
5 |
1 |
59 |
429.54 |
3 |
↓
|
|