|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.28 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
1600 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
6.16 |
-16.49 |
2 |
8 |
0 |
112 |
386.415 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.52 |
5.78 |
-12.54 |
1 |
8 |
0 |
111 |
385.407 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.52 |
5.66 |
-13.46 |
1 |
8 |
0 |
111 |
385.407 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
-1.46 |
-19.34 |
2 |
7 |
0 |
119 |
284.251 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.58 |
-4.86 |
-56.09 |
4 |
7 |
0 |
128 |
285.259 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.85 |
0.21 |
-15.83 |
3 |
7 |
0 |
116 |
285.259 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.26 |
4.03 |
-40.8 |
3 |
8 |
0 |
125 |
402.41 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.62 |
1.56 |
-6.73 |
2 |
4 |
0 |
70 |
202.213 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.62 |
1.7 |
-8.4 |
1 |
4 |
0 |
68 |
201.205 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.29 |
0.67 |
-34.55 |
4 |
4 |
1 |
75 |
203.221 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.62 |
4.99 |
-11.67 |
1 |
7 |
0 |
88 |
361.373 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
3.49 |
1.72 |
-14.32 |
2 |
7 |
0 |
95 |
361.373 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.49 |
2.52 |
-37.73 |
1 |
7 |
-1 |
98 |
360.365 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.38 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
2.83 |
-20.43 |
2 |
8 |
0 |
112 |
423.347 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.88 |
4.1 |
-35.16 |
1 |
8 |
0 |
115 |
422.339 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.88 |
3.76 |
-32.76 |
1 |
8 |
0 |
115 |
422.339 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.36 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
3.58 |
-18.03 |
2 |
8 |
0 |
112 |
437.374 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.77 |
3.52 |
-21.68 |
2 |
8 |
0 |
112 |
437.374 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.32 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
4.28 |
-18.56 |
2 |
8 |
0 |
112 |
437.374 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.15 |
4.4 |
-37.11 |
1 |
8 |
0 |
115 |
436.366 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.15 |
4.77 |
-31.09 |
1 |
8 |
0 |
115 |
436.366 |
7 |
↓
|
|
|
|
Analogs
-
40897533
-
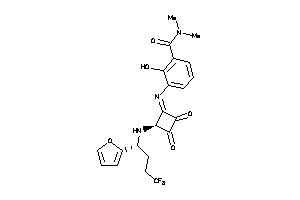
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
4.75 |
-18.83 |
2 |
8 |
0 |
112 |
451.401 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.42 |
5.31 |
-39.24 |
1 |
8 |
0 |
115 |
450.393 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.42 |
5.47 |
-34.83 |
1 |
8 |
0 |
115 |
450.393 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.36 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
3.86 |
-19.12 |
2 |
8 |
0 |
112 |
415.421 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.51 |
3.81 |
-23.62 |
2 |
8 |
0 |
112 |
415.421 |
6 |
↓
|
|
|
|
Analogs
-
40915321
-
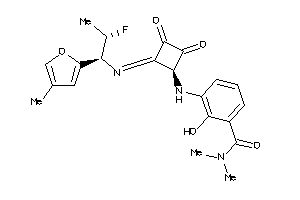
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.39 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.66 |
4.1 |
-19.22 |
2 |
8 |
0 |
112 |
415.421 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.66 |
4.05 |
-18.95 |
2 |
8 |
0 |
112 |
415.421 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.36 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
3.04 |
-19.07 |
2 |
8 |
0 |
112 |
401.394 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.29 |
2.99 |
-18.91 |
2 |
8 |
0 |
112 |
401.394 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.86 |
3.89 |
-19.93 |
2 |
8 |
0 |
112 |
433.411 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.86 |
3.83 |
-22.64 |
2 |
8 |
0 |
112 |
433.411 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
4.13 |
-19.98 |
2 |
8 |
0 |
112 |
433.411 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.02 |
4.07 |
-22.38 |
2 |
8 |
0 |
112 |
433.411 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.34 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
4.04 |
-19.84 |
2 |
8 |
0 |
112 |
433.411 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.02 |
3.99 |
-22.82 |
2 |
8 |
0 |
112 |
433.411 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.32 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.61 |
5.32 |
-19.34 |
2 |
8 |
0 |
112 |
461.465 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.61 |
5.27 |
-21.78 |
2 |
8 |
0 |
112 |
461.465 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.34 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
4.62 |
-19.8 |
2 |
8 |
0 |
112 |
447.438 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.37 |
4.56 |
-22.57 |
2 |
8 |
0 |
112 |
447.438 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.34 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
4.9 |
-19.34 |
2 |
8 |
0 |
112 |
429.448 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.95 |
4.85 |
-18.81 |
2 |
8 |
0 |
112 |
429.448 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[[3,4-dioxo-2-[[(1R)-2,2,3,3,3-pentafluoro-1-(5-methyl-2-furyl)propyl]amino]cyclobuten-1-yl]amino]
3-[[3,4-dioxo-2-[[(1R)-2,2,3,3,3…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.33 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
4.71 |
-17.48 |
2 |
8 |
0 |
112 |
487.381 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
4.66 |
-21.21 |
2 |
8 |
0 |
112 |
487.381 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
34 |
0.34 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
3.45 |
-21.55 |
2 |
8 |
0 |
112 |
457.792 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.68 |
4.72 |
-36.9 |
1 |
8 |
0 |
115 |
456.784 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.68 |
4.39 |
-33.84 |
1 |
8 |
0 |
115 |
456.784 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
103 |
0.31 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
3.71 |
-21.06 |
2 |
8 |
0 |
112 |
492.237 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.29 |
4.92 |
-35.9 |
1 |
8 |
0 |
115 |
491.229 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.29 |
4.6 |
-32.9 |
1 |
8 |
0 |
115 |
491.229 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.35 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.49 |
3.34 |
-20.1 |
2 |
8 |
0 |
112 |
457.792 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.49 |
4.62 |
-34.98 |
1 |
8 |
0 |
115 |
456.784 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.49 |
4.29 |
-32.48 |
1 |
8 |
0 |
115 |
456.784 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.33 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.63 |
4.85 |
-18.44 |
2 |
7 |
0 |
99 |
433.386 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.63 |
5.81 |
-33.73 |
1 |
7 |
0 |
102 |
432.378 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.63 |
5.5 |
-30.37 |
1 |
7 |
0 |
102 |
432.378 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.33 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
3.51 |
-20.98 |
2 |
9 |
0 |
118 |
477.395 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.52 |
4.48 |
-35.5 |
1 |
9 |
0 |
120 |
476.387 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.52 |
4.16 |
-33.51 |
1 |
9 |
0 |
120 |
476.387 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
10.57 |
-57.98 |
1 |
4 |
0 |
51 |
349.454 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.66 |
8.22 |
-9.58 |
0 |
4 |
0 |
50 |
348.446 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.66 |
8.25 |
-7.48 |
0 |
4 |
0 |
50 |
348.446 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
7.39 |
-40.94 |
1 |
4 |
0 |
51 |
285.367 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.40 |
4.91 |
-7.29 |
0 |
4 |
0 |
50 |
284.359 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.40 |
4.94 |
-6.21 |
0 |
4 |
0 |
50 |
284.359 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
9.45 |
-48.01 |
1 |
4 |
0 |
51 |
327.448 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.42 |
9.43 |
-59.46 |
1 |
4 |
0 |
51 |
327.448 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
9.45 |
-47.99 |
1 |
4 |
0 |
51 |
327.448 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.42 |
9.43 |
-59.35 |
1 |
4 |
0 |
51 |
327.448 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
12.38 |
-52.93 |
1 |
4 |
0 |
51 |
389.519 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.59 |
12.27 |
-63.97 |
1 |
4 |
0 |
51 |
389.519 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
9.24 |
-73.41 |
1 |
5 |
0 |
54 |
390.507 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.29 |
9.22 |
-84.73 |
1 |
5 |
0 |
54 |
390.507 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.29 |
6.98 |
-7.75 |
0 |
5 |
0 |
53 |
389.499 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.23 |
8.75 |
-47.37 |
1 |
4 |
0 |
51 |
313.421 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.23 |
8.73 |
-59.44 |
1 |
4 |
0 |
51 |
313.421 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
11.55 |
-59.5 |
1 |
5 |
0 |
54 |
376.48 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.98 |
9.2 |
-7.76 |
0 |
5 |
0 |
53 |
375.472 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
9.93 |
-39.48 |
1 |
4 |
0 |
51 |
341.475 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.47 |
7.83 |
-6.52 |
0 |
4 |
0 |
50 |
340.467 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.47 |
7.85 |
-5.71 |
0 |
4 |
0 |
50 |
340.467 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
9.79 |
-38.61 |
1 |
4 |
0 |
51 |
341.475 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.30 |
7.65 |
-6.28 |
0 |
4 |
0 |
50 |
340.467 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.30 |
7.68 |
-6.14 |
0 |
4 |
0 |
50 |
340.467 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
9.8 |
-37.61 |
1 |
4 |
0 |
51 |
341.475 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.30 |
7.66 |
-6.73 |
0 |
4 |
0 |
50 |
340.467 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.30 |
7.69 |
-5.75 |
0 |
4 |
0 |
50 |
340.467 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
10.67 |
-47.5 |
1 |
4 |
0 |
51 |
355.502 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.48 |
10.64 |
-59.13 |
1 |
4 |
0 |
51 |
355.502 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
10.67 |
-47.52 |
1 |
4 |
0 |
51 |
355.502 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.48 |
10.64 |
-59.27 |
1 |
4 |
0 |
51 |
355.502 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
10.49 |
-73.23 |
1 |
5 |
0 |
54 |
418.561 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.35 |
10.47 |
-84.74 |
1 |
5 |
0 |
54 |
418.561 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.35 |
8.23 |
-7.5 |
0 |
5 |
0 |
53 |
417.553 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.13 |
10.71 |
-53.81 |
1 |
5 |
0 |
54 |
424.609 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.13 |
8.5 |
-6.36 |
0 |
5 |
0 |
53 |
423.601 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.13 |
8.47 |
-7.91 |
0 |
5 |
0 |
53 |
423.601 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
12.08 |
-52.49 |
1 |
4 |
0 |
51 |
389.519 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.06 |
9.84 |
-7.06 |
0 |
4 |
0 |
50 |
388.511 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.06 |
9.79 |
-8.57 |
0 |
4 |
0 |
50 |
388.511 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
11.98 |
-16.91 |
0 |
9 |
0 |
119 |
524.51 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
10.28 |
-12.95 |
0 |
6 |
0 |
91 |
401.466 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.98 |
9.91 |
-11.18 |
0 |
6 |
0 |
91 |
401.466 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
10.7 |
-13.2 |
0 |
6 |
0 |
91 |
401.466 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.98 |
9.52 |
-11.67 |
0 |
6 |
0 |
91 |
401.466 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
13.11 |
-15.58 |
0 |
6 |
0 |
91 |
427.504 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.62 |
13.1 |
-17.19 |
0 |
6 |
0 |
91 |
427.504 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
7.25 |
-10.69 |
0 |
6 |
0 |
91 |
339.395 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.31 |
6.93 |
-13.71 |
0 |
6 |
0 |
91 |
339.395 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
7.09 |
-12.36 |
0 |
6 |
0 |
91 |
339.395 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.31 |
7.11 |
-14.04 |
0 |
6 |
0 |
91 |
339.395 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.05 |
5.33 |
-11.54 |
0 |
6 |
0 |
91 |
297.314 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
7.1 |
-12.26 |
0 |
6 |
0 |
91 |
339.395 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
7.1 |
-12.51 |
0 |
6 |
0 |
91 |
339.395 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.36 |
3.34 |
-9.54 |
0 |
6 |
0 |
80 |
287.319 |
3 |
↓
|
|