|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
5.27 |
-6.86 |
1 |
3 |
0 |
47 |
228.295 |
1 |
↓
|
|
|
|
Analogs
-
3940618
-
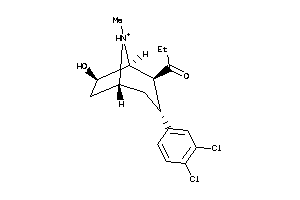
-
3940615
-
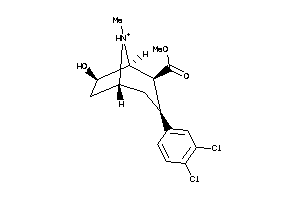
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
SC6A4-4-E |
Serotonin Transporter (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1220 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.52 |
6.84 |
-46.23 |
2 |
4 |
1 |
51 |
345.246 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.52 |
4.51 |
-7.6 |
1 |
4 |
0 |
50 |
344.238 |
3 |
↓
|
|
|
|
Analogs
-
13742498
-
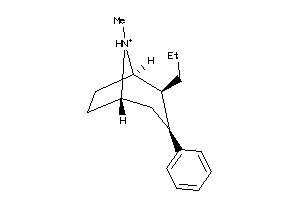
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
95 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.73 |
10.43 |
-34.32 |
1 |
1 |
1 |
4 |
244.402 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.61 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
24 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
10.5 |
-38.06 |
1 |
1 |
1 |
4 |
262.392 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.68 |
11.89 |
-34.54 |
1 |
1 |
1 |
4 |
272.456 |
4 |
↓
|
|
|
|
Analogs
-
36177169
-
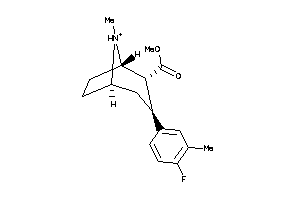
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
8.94 |
-44.01 |
1 |
3 |
1 |
31 |
292.374 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.69 |
6.6 |
-7.61 |
0 |
3 |
0 |
30 |
291.366 |
3 |
↓
|
|
|
|
Analogs
-
13738423
-
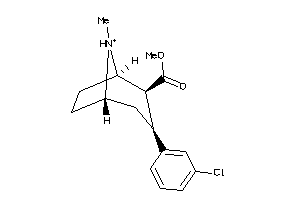
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
8.97 |
-32.46 |
1 |
3 |
1 |
31 |
294.802 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.81 |
7.63 |
-6.51 |
0 |
3 |
0 |
30 |
293.794 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
7.03 |
-32.09 |
3 |
4 |
1 |
57 |
354.268 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.37 |
5.69 |
-6.78 |
2 |
4 |
0 |
56 |
353.26 |
3 |
↓
|
|
|
|
Analogs
-
26259313
-
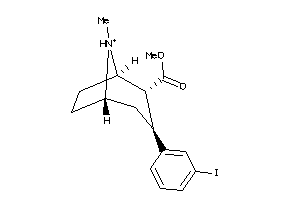
-
13738424
-
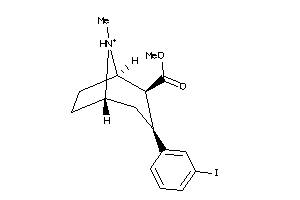
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.21 |
9.53 |
-32.5 |
1 |
3 |
1 |
31 |
386.253 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.21 |
8.19 |
-6.3 |
0 |
3 |
0 |
30 |
385.245 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
7.5 |
-31.98 |
3 |
4 |
1 |
57 |
401.268 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.65 |
6.16 |
-6.57 |
2 |
4 |
0 |
56 |
400.26 |
3 |
↓
|
|
|
|
Analogs
-
13738422
-
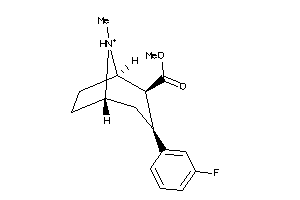
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
8.52 |
-33.16 |
1 |
3 |
1 |
31 |
278.347 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.29 |
7.18 |
-7.19 |
0 |
3 |
0 |
30 |
277.339 |
3 |
↓
|
|
|
|
Analogs
-
13738421
-
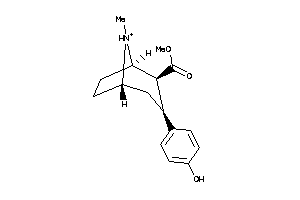
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
5.59 |
-33.16 |
2 |
4 |
1 |
51 |
276.356 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.67 |
4.26 |
-8.03 |
1 |
4 |
0 |
50 |
275.348 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
12.37 |
-39.72 |
1 |
3 |
1 |
31 |
426.318 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.13 |
10.17 |
-3.74 |
0 |
3 |
0 |
30 |
425.31 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
10.16 |
-8.86 |
0 |
3 |
0 |
30 |
329.415 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.50 |
11.41 |
-34.12 |
1 |
3 |
1 |
31 |
330.423 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
87 |
0.41 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.78 |
11.98 |
-38.72 |
1 |
3 |
1 |
31 |
397.308 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.78 |
11 |
-8.07 |
0 |
3 |
0 |
30 |
396.3 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
9.38 |
-38.82 |
1 |
2 |
1 |
22 |
272.412 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
145 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.05 |
12.46 |
-38.39 |
1 |
3 |
1 |
31 |
444.308 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.05 |
11.47 |
-7.87 |
0 |
3 |
0 |
30 |
443.3 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
9.08 |
-43.08 |
1 |
9 |
1 |
122 |
350.351 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.52 |
6.72 |
-5.63 |
0 |
9 |
0 |
121 |
349.343 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.13 |
4.46 |
-36.07 |
3 |
2 |
1 |
37 |
240.273 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
9.41 |
-45.6 |
2 |
4 |
1 |
51 |
356.486 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.77 |
7.05 |
-10.35 |
1 |
4 |
0 |
50 |
355.478 |
6 |
↓
|
|
|
|
Analogs
-
36177332
-
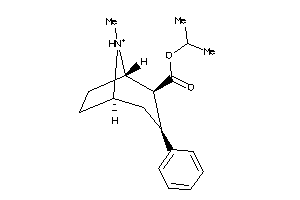
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
10.25 |
-38.21 |
1 |
3 |
1 |
31 |
302.438 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.34 |
8.19 |
-6.11 |
0 |
3 |
0 |
30 |
301.43 |
4 |
↓
|
|
|
|
Analogs
-
6522704
-
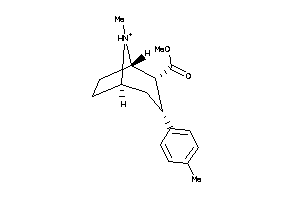
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.07 |
9.67 |
-38.87 |
1 |
3 |
1 |
31 |
288.411 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.07 |
7.32 |
-6.26 |
0 |
3 |
0 |
30 |
287.403 |
4 |
↓
|
|