|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
95 |
0.55 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1300 |
0.46 |
Binding ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADCY3-1-E |
Adenylate Cyclase Type III (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADCY4-1-E |
Adenylate Cyclase Type IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADCY5-1-E |
Adenylate Cyclase Type V (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADCY6-1-E |
Adenylate Cyclase Type VI (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADCY8-2-E |
Adenylate Cyclase Type VIII (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.48 |
5.57 |
-7.83 |
1 |
6 |
0 |
73 |
248.286 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
585 |
0.51 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
922 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
5.85 |
-19.41 |
2 |
5 |
0 |
70 |
225.255 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.40 |
Binding ≤ 10μM
|
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
158 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.29 |
5.76 |
-13.58 |
1 |
6 |
0 |
69 |
361.401 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
4.29 |
6.15 |
-55 |
0 |
6 |
-1 |
72 |
360.393 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.29 |
7.08 |
-18.69 |
1 |
6 |
0 |
73 |
361.401 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
540 |
0.35 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
530 |
0.35 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.42 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
42 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
10.13 |
-45.92 |
5 |
9 |
1 |
129 |
338.351 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.12 |
9.61 |
-15.7 |
4 |
9 |
0 |
127 |
337.343 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
4.03 |
-15.89 |
4 |
10 |
0 |
134 |
510.292 |
5 |
↓
|
|
|
|
Analogs
-
16603751
-
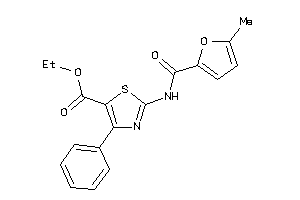
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
95 |
0.41 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
13 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
7.86 |
-10.39 |
1 |
6 |
0 |
81 |
342.376 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.42 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
AA1R-3-E |
Adenosine A1 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
980 |
0.38 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2800 |
0.35 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1400 |
0.37 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4500 |
0.34 |
Binding ≤ 10μM
|
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.49 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
65 |
0.46 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.47 |
Binding ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
530 |
0.40 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
840 |
0.39 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
AOFA-1-E |
Monoamine Oxidase A (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
GCR-1-E |
Glucocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
610 |
0.40 |
Binding ≤ 10μM
|
|
HRH2-3-E |
Histamine H2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
HRH3-3-E |
Histamine H3 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
MC4R-1-E |
Melanocortin Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
MK14-2-E |
MAP Kinase P38 Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
MT3-1-E |
Metallothionein-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
NISCH-2-E |
Nischarin (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
NK1R-1-E |
Neurokinin 1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
NK2R-1-E |
Neurokinin 2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
NPY1R-2-E |
Neuropeptide Y Receptor Type 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
NTR1-1-E |
Neurotensin Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
OPRK-4-E |
Kappa Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
SC5A7-2-E |
High Affinity Choline Transporter 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.43 |
Functional ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Binding ≤ 10μM
|
|
Z104283-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #2 Of 4), Other |
Other |
1000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.33 |
2.67 |
-39.39 |
1 |
2 |
1 |
21 |
302.482 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1111 |
0.29 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
10.9 |
-14.99 |
2 |
9 |
0 |
109 |
389.419 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
220 |
0.33 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
884 |
0.30 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.35 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.33 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
53 |
0.36 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
96 |
0.35 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.30 |
Functional ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1680 |
0.29 |
Functional ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.30 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 5), Other |
Other |
900 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
2.3 |
-16.14 |
4 |
9 |
0 |
126 |
385.424 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
210 |
0.55 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.45 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6600 |
0.43 |
Binding ≤ 10μM
|
|
ADCY1-1-E |
Brain Adenylate Cyclase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
|
ADCY3-1-E |
Adenylate Cyclase Type III (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
|
ADCY5-1-E |
Adenylate Cyclase Type V (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
|
ADCY6-1-E |
Adenylate Cyclase Type VI (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
|
ADCY7-1-E |
Adenylate Cyclase Type VII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
|
ADCY8-2-E |
Adenylate Cyclase Type VIII (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
|
ADCY9-1-E |
Adenylate Cyclase Type IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.46 |
Functional ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.51 |
Functional ≤ 10μM
|
|
Z50512-5-O |
Cavia Porcellus (cluster #5 Of 7), Other |
Other |
8100 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
0.06 |
-10.94 |
1 |
6 |
0 |
72 |
236.275 |
4 |
↓
|
|
|
|
Analogs
-
39851135
-
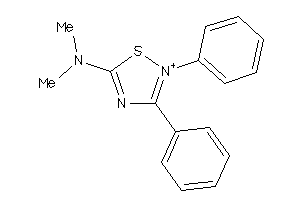
-
39851136
-
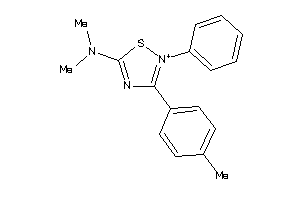
-
39851137
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2800 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.46 |
8.95 |
-21.66 |
1 |
3 |
1 |
29 |
268.365 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.46 |
9.44 |
-84.96 |
2 |
3 |
2 |
30 |
269.373 |
3 |
↓
|
|
|
|
Analogs
-
21792752
-
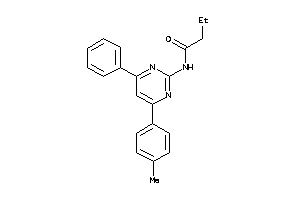
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3760 |
0.29 |
Binding ≤ 10μM
|
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
131 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.83 |
13.59 |
-15.13 |
1 |
4 |
0 |
55 |
345.446 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
7.88 |
-10.35 |
2 |
6 |
0 |
82 |
285.694 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
410 |
0.37 |
Binding ≤ 10μM |
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.48 |
Binding ≤ 10μM |
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
260 |
0.38 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
6.91 |
-16.47 |
2 |
5 |
0 |
71 |
353.394 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
10.81 |
-11.79 |
2 |
8 |
0 |
100 |
345.366 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
2.24 |
-14.07 |
0 |
8 |
0 |
80 |
412.49 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.40 |
2.5 |
-45.67 |
1 |
8 |
1 |
82 |
413.498 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
95 |
0.45 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
706 |
0.39 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
590 |
0.40 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
795 |
0.39 |
Binding ≤ 10μM
|
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Functional ≤ 10μM |
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
69 |
0.46 |
Functional ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
69 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
8.99 |
-7.17 |
1 |
6 |
0 |
73 |
304.394 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
-10.66 |
-15.83 |
4 |
10 |
0 |
134 |
544.737 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 78 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9800 |
0.54 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9400 |
0.54 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.54 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
14 |
0.85 |
Binding ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY3-1-E |
Adenylate Cyclase Type III (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY4-1-E |
Adenylate Cyclase Type IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY5-1-E |
Adenylate Cyclase Type V (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY6-1-E |
Adenylate Cyclase Type VI (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY8-2-E |
Adenylate Cyclase Type VIII (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Functional ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 5), Other |
Other |
9000 |
0.54 |
Binding ≤ 10μM
|
|
Z50512-5-O |
Cavia Porcellus (cluster #5 Of 7), Other |
Other |
6520 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.00 |
3.24 |
-11.33 |
1 |
6 |
0 |
73 |
180.167 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
72 |
0.50 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.57 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
352 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
-1.46 |
-14.62 |
2 |
8 |
0 |
100 |
272.316 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.78 |
-1.35 |
-33.28 |
3 |
8 |
1 |
102 |
273.324 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4600 |
0.39 |
Binding ≤ 10μM
|
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6800 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
8 |
-6.42 |
2 |
2 |
0 |
43 |
291.803 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.52 |
7.32 |
-14.24 |
2 |
10 |
0 |
124 |
508.604 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.04 |
6.07 |
-41.35 |
1 |
10 |
-1 |
127 |
507.596 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.52 |
9.54 |
-56.24 |
3 |
10 |
1 |
125 |
509.612 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
865 |
0.45 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
106 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
7.28 |
-7.24 |
2 |
3 |
0 |
63 |
253.28 |
2 |
↓
|
|
|
|
Analogs
-
34944216
-
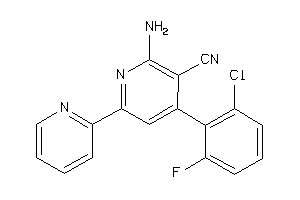
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
239 |
0.42 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
89 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.47 |
9.23 |
-8.75 |
2 |
3 |
0 |
63 |
305.768 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
28 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.44 |
13.08 |
-13.93 |
1 |
4 |
0 |
55 |
331.419 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
309 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.21 |
13.84 |
-14.84 |
1 |
4 |
0 |
55 |
351.409 |
4 |
↓
|
|
|
|
Analogs
-
34427996
-
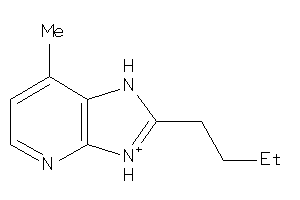
-
34435464
-
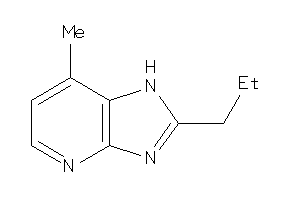
-
38482185
-
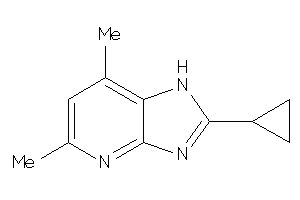
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
101 |
0.75 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
5.14 |
-15.56 |
1 |
3 |
0 |
42 |
175.235 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.01 |
5.51 |
-28.12 |
2 |
3 |
1 |
43 |
176.243 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
119 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.78 |
14.04 |
-14.48 |
1 |
4 |
0 |
55 |
357.457 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
42 |
0.40 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
267 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
4.11 |
-11.21 |
4 |
6 |
0 |
112 |
347.374 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.13 |
5.65 |
-48.79 |
4 |
6 |
0 |
112 |
347.374 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.93 |
0.62 |
-13.94 |
1 |
4 |
0 |
54 |
317.392 |
5 |
↓
|
|
|
|
Analogs
-
34953724
-

-
19841853
-
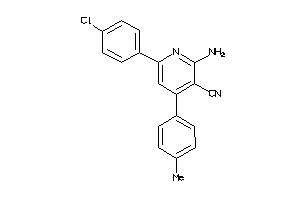
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
192 |
0.43 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
201 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
9.35 |
-7.31 |
2 |
3 |
0 |
63 |
305.768 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
224 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.20 |
12.74 |
-13.57 |
1 |
4 |
0 |
55 |
331.419 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
203 |
0.47 |
Binding ≤ 10μM |
|
AA2AR-3-E |
Adenosine A2a Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8600 |
0.35 |
Binding ≤ 10μM
|
|
AA3R-4-E |
Adenosine Receptor A3 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
31 |
0.53 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
7.17 |
-15.95 |
1 |
4 |
0 |
51 |
261.284 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.30 |
5.1 |
-62.41 |
0 |
4 |
-1 |
54 |
260.276 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
270 |
0.34 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.39 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
35 |
0.39 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
660 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
7.23 |
-47.09 |
2 |
9 |
1 |
94 |
368.421 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.35 |
4.84 |
-18.78 |
1 |
9 |
0 |
92 |
367.413 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1730 |
0.28 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
14 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
8.67 |
-46.92 |
2 |
9 |
1 |
94 |
396.475 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.79 |
6.28 |
-18.72 |
1 |
9 |
0 |
92 |
395.467 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
647 |
0.58 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
48 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.64 |
3.23 |
-10.85 |
3 |
6 |
0 |
94 |
201.189 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
11 |
0.53 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
19 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
8.26 |
-6.78 |
2 |
4 |
0 |
76 |
272.311 |
2 |
↓
|
|
|
|
Analogs
-
39283389
-
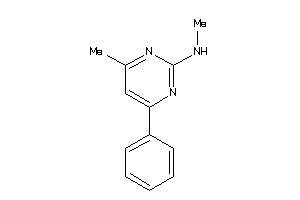
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-3-E |
Adenosine A1 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
18 |
0.57 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
405 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
9.19 |
-8.41 |
2 |
3 |
0 |
52 |
247.301 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
6.64 |
-62.62 |
5 |
10 |
1 |
139 |
429.501 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.35 |
5.37 |
-83.46 |
4 |
10 |
0 |
142 |
428.493 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.84 |
6.26 |
-17.96 |
4 |
10 |
0 |
137 |
428.493 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
12.48 |
-21.39 |
2 |
10 |
0 |
135 |
486.532 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.46 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.45 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9100 |
0.44 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
800 |
0.53 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.48 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.48 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.48 |
Binding ≤ 10μM
|
|
PDE3A-1-E |
Phosphodiesterase 3A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE3B-1-E |
Phosphodiesterase 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE4A-3-E |
Phosphodiesterase 4A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE4B-2-E |
Phosphodiesterase 4B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE4C-2-E |
Phosphodiesterase 4C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE4D-2-E |
Phosphodiesterase 4D (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3020 |
0.48 |
Binding ≤ 10μM
|
|
PDE7A-1-E |
Phosphodiesterase 7A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.47 |
Binding ≤ 10μM
|
|
PDE7B-1-E |
Phosphodiesterase 7B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.47 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.50 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.50 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.50 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.50 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.45 |
Functional ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.46 |
Functional ≤ 10μM
|
|
PDE1B-2-E |
Phosphodiesterase 1B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.61 |
Functional ≤ 10μM
|
|
PDE1C-2-E |
Phosphodiesterase 1C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.61 |
Functional ≤ 10μM
|
|
Q9EPR9-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.61 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 5), Other |
Other |
800 |
0.53 |
Binding ≤ 10μM
|
|
Z50588-1-O |
Canis Familiaris (cluster #1 Of 7), Other |
Other |
5500 |
0.46 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3500 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
5.25 |
-10.93 |
1 |
6 |
0 |
73 |
222.248 |
2 |
↓
|
|
|
|
Analogs
-
39125283
-
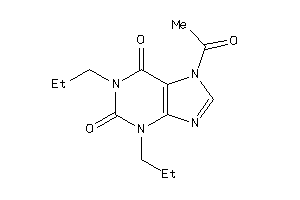
-
39373696
-
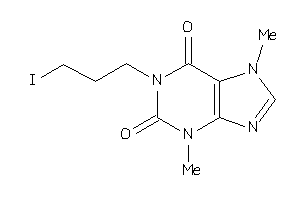
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7700 |
0.40 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5300 |
0.41 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5300 |
0.41 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
8.34 |
-10.87 |
0 |
6 |
0 |
62 |
250.302 |
4 |
↓
|
|
|
|
Analogs
-
4475124
-
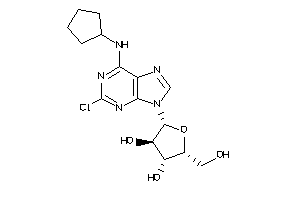
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
-0.44 |
-13.95 |
4 |
9 |
0 |
126 |
369.809 |
4 |
↓
|
|
|
|
Analogs
-
4475112
-
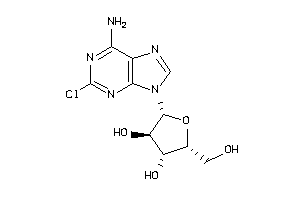
-
4536052
-
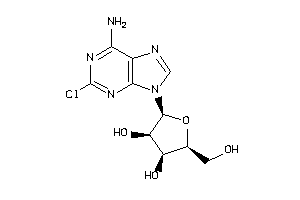
-
4536053
-
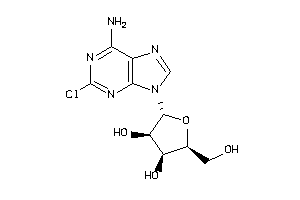
-
4536054
-
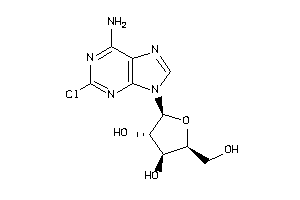
-
4536057
-
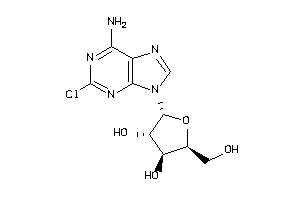
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
18 |
0.54 |
Binding ≤ 10μM |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
20 |
0.54 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
76 |
0.50 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
920 |
0.42 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.49 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3200 |
0.38 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Functional ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.44 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 5), Other |
Other |
920 |
0.42 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
7943 |
0.36 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
620 |
0.43 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
10000 |
0.35 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
3000 |
0.39 |
Functional ≤ 10μM
|
|
Z80945-1-O |
HEp-2 (Laryngeal Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
3000 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.14 |
-4.58 |
-14.68 |
5 |
9 |
0 |
140 |
301.69 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.87 |
4.92 |
-11.58 |
1 |
6 |
0 |
73 |
208.221 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
6.05 |
-7.92 |
2 |
5 |
0 |
70 |
203.249 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
-0.19 |
-22.78 |
2 |
11 |
0 |
124 |
432.444 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6900 |
0.34 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
7.73 |
-9.52 |
2 |
5 |
0 |
69 |
295.733 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
4475123
-
-
3872014
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.61 |
6.05 |
-59.85 |
6 |
13 |
-1 |
201 |
498.52 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.42 |
7.24 |
-6.72 |
0 |
1 |
0 |
17 |
194.299 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.61 |
6.43 |
-70.08 |
7 |
13 |
0 |
202 |
499.528 |
10 |
↓
|
|