|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
4.23 |
-7.59 |
0 |
3 |
0 |
47 |
228.034 |
0 |
↓
|
|
|
|
Analogs
-
5724366
-
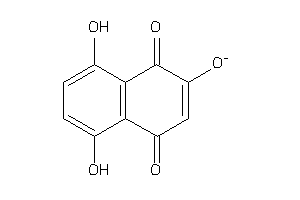
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.13 |
2.17 |
-9.54 |
2 |
4 |
0 |
75 |
190.154 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
519 |
0.73 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.07 |
0.32 |
-9.94 |
0 |
3 |
0 |
47 |
159.144 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
0.2 |
-4.84 |
0 |
3 |
0 |
47 |
228.034 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.30 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
640 |
0.36 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
3.23 |
-41.73 |
1 |
5 |
-1 |
86 |
451.094 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.25 |
3.99 |
-92.12 |
0 |
5 |
-2 |
88 |
450.086 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.28 |
5.32 |
-46.11 |
0 |
5 |
-1 |
82 |
451.094 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.48 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
95 |
0.45 |
Binding ≤ 10μM
|
|
MPIP3-2-E |
Dual Specificity Phosphatase Cdc25C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
89 |
0.45 |
Binding ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
2300 |
0.36 |
Functional ≤ 10μM
|
|
Z80800-1-O |
MIA PaCa-2 (Pancreatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2500 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
1.28 |
-9.59 |
0 |
6 |
0 |
72 |
321.764 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.42 |
1.17 |
-10.76 |
0 |
6 |
0 |
72 |
321.764 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.42 |
3.55 |
-37.71 |
1 |
6 |
1 |
73 |
322.772 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3280 |
0.28 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4670 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.97 |
7.88 |
-14.34 |
2 |
6 |
0 |
88 |
401.875 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.97 |
8.81 |
-47.57 |
1 |
6 |
-1 |
91 |
400.867 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.37 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3770 |
0.38 |
Binding ≤ 10μM
|
|
MPIP3-2-E |
Dual Specificity Phosphatase Cdc25C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
57 |
0.51 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.39 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
8500 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.13 |
2.29 |
-10.08 |
2 |
4 |
0 |
75 |
310.396 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
3.54 |
-8.75 |
1 |
3 |
0 |
54 |
174.155 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.40 |
4.54 |
-46.63 |
0 |
3 |
-1 |
57 |
173.147 |
0 |
↓
|
|
|
|
Analogs
-
1237523
-
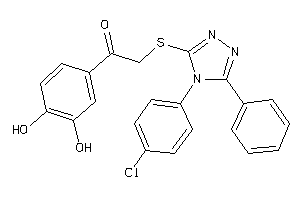
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.27 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.07 |
-1.08 |
-14.04 |
2 |
6 |
0 |
88 |
451.935 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1750 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
7.02 |
-112.56 |
0 |
6 |
-2 |
114 |
336.346 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4550 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
7.49 |
-50.03 |
0 |
4 |
-1 |
74 |
275.305 |
4 |
↓
|
|
|
|
Analogs
-
2163434
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-2-E |
Dual Specificity Protein Phosphatase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.57 |
Binding ≤ 10μM
|
|
DUS6-1-E |
Dual Specificity Protein Phosphatase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1350 |
0.55 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
270 |
0.61 |
Binding ≤ 10μM
|
|
PTPRC-3-E |
Leukocyte Common Antigen (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.53 |
Binding ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
370 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
1.93 |
-10.73 |
0 |
3 |
0 |
43 |
202.209 |
2 |
↓
|
|
|
|
Analogs
-
39866685
-
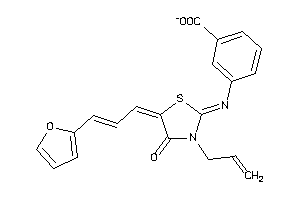
-
5376740
-
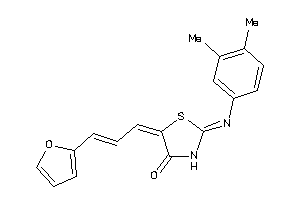
-
5376748
-
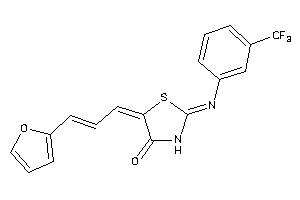
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7100 |
0.31 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5440 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.30 |
9.26 |
-11.17 |
1 |
4 |
0 |
58 |
324.405 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.81 |
8.79 |
-48.19 |
0 |
4 |
-1 |
57 |
323.397 |
4 |
↓
|
|
|
|
Analogs
-
5376767
-
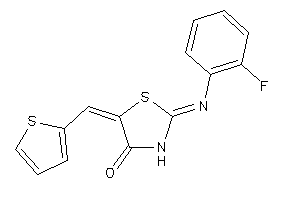
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8900 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.21 |
7.01 |
-37.47 |
0 |
3 |
-1 |
44 |
303.363 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
4.70 |
7.56 |
-6.83 |
1 |
3 |
0 |
45 |
304.371 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.16 |
5.08 |
-43.24 |
0 |
3 |
-1 |
48 |
303.363 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.33 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.27 |
9.05 |
-15.45 |
1 |
4 |
0 |
55 |
365.241 |
4 |
↓
|
|
|
|
Analogs
-
16666712
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.58 |
8.87 |
-38.34 |
1 |
4 |
-1 |
60 |
387.271 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.59 |
9.48 |
-15.08 |
2 |
4 |
0 |
58 |
388.279 |
3 |
↓
|
|
|
|
Analogs
-
5376752
-
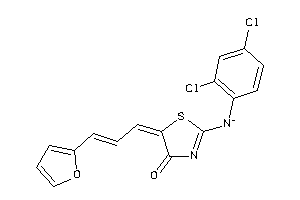
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6700 |
0.31 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.27 |
8.7 |
-39.95 |
0 |
4 |
-1 |
57 |
364.233 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
5.76 |
9.08 |
-9.84 |
1 |
4 |
0 |
58 |
365.241 |
3 |
↓
|
|
|
|
Analogs
-
1260961
-
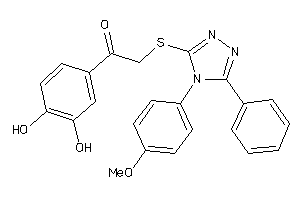
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9200 |
0.22 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
-0.92 |
-15.63 |
2 |
7 |
0 |
97 |
447.516 |
7 |
↓
|
|
|
|
Analogs
-
2596377
-
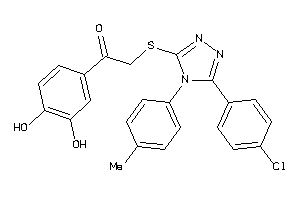
-
1260822
-
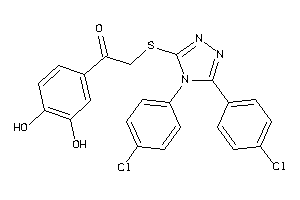
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.63 |
-1.41 |
-14.35 |
2 |
6 |
0 |
88 |
437.908 |
6 |
↓
|
|
|
|
Analogs
-
16669657
-
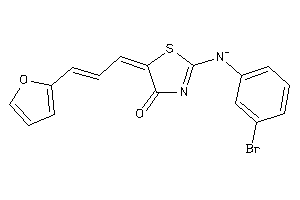
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7700 |
0.33 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
8.18 |
-42.18 |
0 |
4 |
-1 |
57 |
374.239 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
2 |
-29.71 |
1 |
9 |
0 |
122 |
491.504 |
8 |
↓
|
|
|
|
Analogs
-
19911459
-
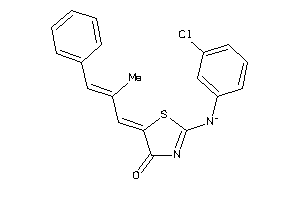
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.11 |
10.23 |
-43.77 |
0 |
3 |
-1 |
44 |
353.854 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.11 |
10.65 |
-19.18 |
1 |
3 |
0 |
42 |
354.862 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8100 |
0.29 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.28 |
0.32 |
-14.93 |
1 |
3 |
0 |
41 |
348.471 |
4 |
↓
|
|
|
|
Analogs
-
39869651
-
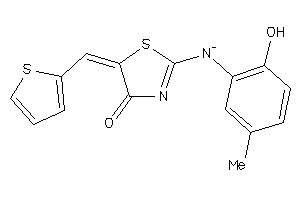
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7800 |
0.36 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
4.62 |
-8.98 |
2 |
4 |
0 |
65 |
302.38 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
3.55 |
4.09 |
-40.91 |
1 |
4 |
-1 |
64 |
301.372 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.50 |
2.83 |
-46.44 |
1 |
4 |
-1 |
69 |
301.372 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3900 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.08 |
0.02 |
-10.98 |
1 |
3 |
0 |
41 |
354.862 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5100 |
0.30 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.74 |
-0.65 |
-13.84 |
1 |
3 |
0 |
41 |
389.307 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9510 |
0.44 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM |
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
580 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.67 |
7.32 |
-7.86 |
0 |
2 |
0 |
34 |
208.216 |
0 |
↓
|
|
|
|
Analogs
-
1276363
-
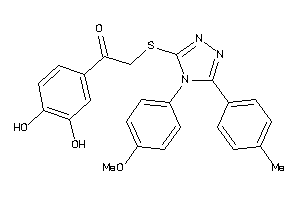
-
1287774
-
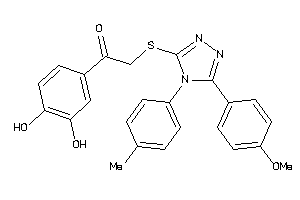
-
1260961
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.24 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
-1.23 |
-15.98 |
2 |
7 |
0 |
97 |
433.489 |
7 |
↓
|
|
|
|
Analogs
-
5376627
-

-
5376628
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.08 |
9.27 |
-7.96 |
2 |
4 |
0 |
61 |
388.279 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
5.58 |
8.74 |
-38.89 |
1 |
4 |
-1 |
60 |
387.271 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.24 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
10.84 |
-54.49 |
0 |
7 |
-1 |
103 |
438.492 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.28 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9100 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
-1.14 |
-19.16 |
2 |
7 |
0 |
99 |
343.346 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.15 |
1.33 |
-13.34 |
0 |
5 |
0 |
60 |
386.48 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.26 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
210 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.80 |
10.34 |
-52.33 |
0 |
6 |
-1 |
79 |
413.482 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3900 |
0.33 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
8.44 |
-6.71 |
0 |
3 |
0 |
47 |
329.758 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.29 |
8.34 |
-8.39 |
0 |
3 |
0 |
47 |
329.758 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
1.94 |
-15.29 |
0 |
7 |
0 |
93 |
388.836 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-2-E |
Dual Specificity Protein Phosphatase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8450 |
0.59 |
Binding ≤ 10μM
|
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
990 |
0.70 |
Binding ≤ 10μM
|
|
MP2K1-3-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
380 |
0.75 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2760 |
0.65 |
Binding ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
127 |
0.80 |
Functional ≤ 10μM
|
|
Z81034-10-O |
A2780 (Ovarian Carcinoma Cells) (cluster #10 Of 10), Other |
Other |
2300 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
5.04 |
-6.65 |
0 |
2 |
0 |
34 |
158.156 |
0 |
↓
|
|
|
|
Analogs
-
1874920
-
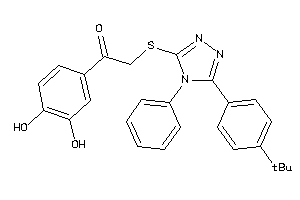
-
5987855
-
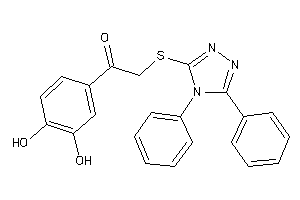
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7100 |
0.24 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.39 |
10.03 |
-15.83 |
2 |
6 |
0 |
88 |
417.49 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
12.75 |
-13.2 |
0 |
5 |
0 |
61 |
375.453 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
0.89 |
-12.52 |
0 |
5 |
0 |
60 |
358.854 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8600 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
9.35 |
-47.84 |
0 |
4 |
-1 |
74 |
277.255 |
2 |
↓
|
|
|
|
Analogs
-
39864224
-
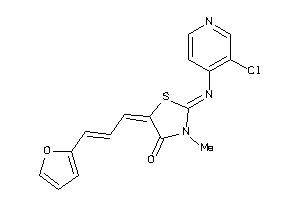
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9100 |
0.32 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
8.55 |
-10.74 |
1 |
4 |
0 |
58 |
330.796 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
4.64 |
8.08 |
-42.21 |
0 |
4 |
-1 |
57 |
329.788 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5100 |
0.19 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.24 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8700 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.48 |
3.16 |
-41.83 |
4 |
12 |
-1 |
190 |
519.49 |
7 |
↓
|
|
|
|
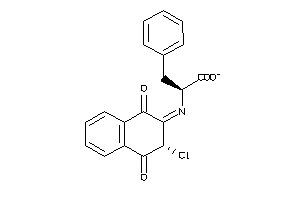
Analogs
-
4937944
-
-
4937948
-
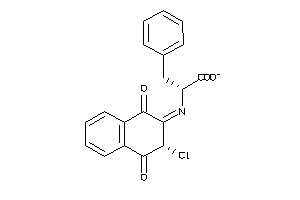
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.31 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
0.91 |
9.18 |
-55.16 |
0 |
5 |
-1 |
87 |
354.769 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
5376750
-
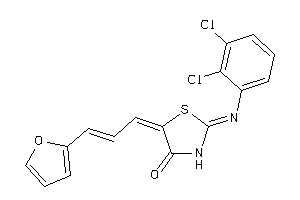
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.32 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.25 |
8.65 |
-41.45 |
0 |
4 |
-1 |
57 |
364.233 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
5.74 |
9.16 |
-11.63 |
1 |
4 |
0 |
58 |
365.241 |
3 |
↓
|
|
|
|
Analogs
-
3063275
-
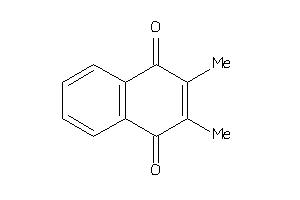
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
5.73 |
-6.36 |
0 |
2 |
0 |
34 |
172.183 |
0 |
↓
|
|