|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R2-2-E |
Prostanoid EP2 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
|
PE2R3-2-E |
Prostanoid EP3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.33 |
Binding ≤ 10μM
|
|
PE2R4-2-E |
Prostanoid EP4 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
8.77 |
-55.02 |
1 |
5 |
-1 |
81 |
340.484 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R2-2-E |
Prostanoid EP2 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
|
PE2R3-2-E |
Prostanoid EP3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.33 |
Binding ≤ 10μM
|
|
PE2R4-2-E |
Prostanoid EP4 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
8.72 |
-55.53 |
1 |
5 |
-1 |
81 |
340.484 |
14 |
↓
|
|
|
|
Analogs
-
39592808
-
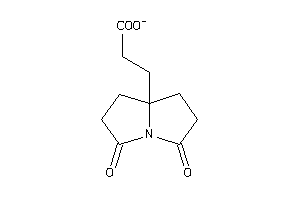
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
8.8 |
-54.94 |
1 |
5 |
-1 |
81 |
340.484 |
14 |
↓
|
|
|
|
Analogs
-
39592808
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
8.87 |
-53.47 |
1 |
5 |
-1 |
81 |
340.484 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
8.06 |
-51.37 |
2 |
5 |
-1 |
98 |
393.544 |
13 |
↓
|
|
|
|
Analogs
-
36878455
-
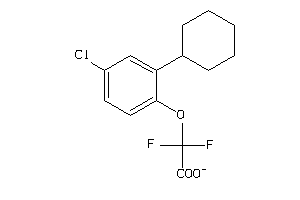
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPR44-1-E |
G Protein-coupled Receptor 44 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
154 |
0.53 |
Binding ≤ 10μM
|
|
PD2R-2-E |
Prostanoid DP Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7600 |
0.40 |
Binding ≤ 10μM
|
|
PE2R2-1-E |
Prostanoid EP2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
1.13 |
-46.07 |
0 |
3 |
-1 |
49 |
267.732 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PD2R-1-E |
Prostanoid DP Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.19 |
Binding ≤ 10μM
|
|
PE2R2-1-E |
Prostanoid EP2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.19 |
Binding ≤ 10μM
|
|
PE2R3-1-E |
Prostanoid EP3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.18 |
Binding ≤ 10μM
|
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.27 |
Binding ≤ 10μM
|
|
PF2R-1-E |
Prostaglandin F2-alpha Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.19 |
Binding ≤ 10μM
|
|
TA2R-1-E |
Thromboxane A2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
630 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.47 |
10.46 |
-61.22 |
0 |
10 |
-1 |
130 |
602.689 |
11 |
↓
|
|
|
|
Analogs
-
27646610
-
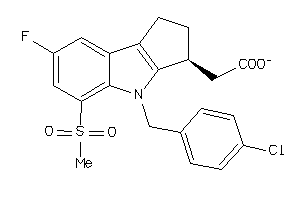
-
34826099
-
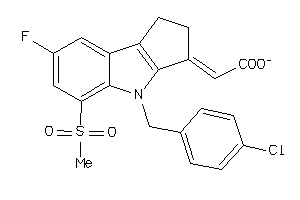
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPR44-1-E |
G Protein-coupled Receptor 44 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
745 |
0.30 |
Binding ≤ 10μM
|
|
PD2R-1-E |
Prostanoid DP Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.43 |
Binding ≤ 10μM
|
|
PD2R-1-E |
Prostanoid DP Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
|
PE2R1-1-E |
Prostanoid EP1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1160 |
0.29 |
Binding ≤ 10μM
|
|
PE2R2-1-E |
Prostanoid EP2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
136 |
0.33 |
Binding ≤ 10μM
|
|
PE2R3-1-E |
Prostanoid EP3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
892 |
0.29 |
Binding ≤ 10μM
|
|
PF2R-1-E |
Prostaglandin F2-alpha Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9991 |
0.24 |
Binding ≤ 10μM
|
|
PI2R-1-E |
Prostanoid IP Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6628 |
0.25 |
Binding ≤ 10μM
|
|
TA2R-1-E |
Thromboxane A2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.41 |
Binding ≤ 10μM
|
|
PD2R-1-E |
Prostanoid DP Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.41 |
Functional ≤ 10μM
|
|
TA2R-1-E |
Thromboxane A2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
770 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
-1.2 |
-48.64 |
0 |
5 |
-1 |
79 |
434.896 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
7.27 |
15.64 |
-7.97 |
0 |
3 |
0 |
36 |
414.545 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.89 |
14.08 |
-59.26 |
1 |
4 |
-1 |
69 |
472.508 |
7 |
↓
|
|