|
|
Analogs
-
2086
-
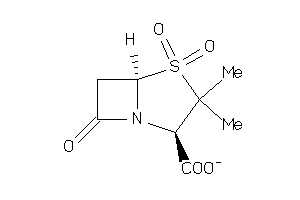
Draw
Identity
99%
90%
80%
70%
Vendors
And 58 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A1E3K9-1-B |
Beta-lactamase SCO-1 (cluster #1 Of 1), Bacterial |
Bacteria |
6200 |
0.49 |
Binding ≤ 10μM
|
|
A8RR46-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
75 |
0.66 |
Binding ≤ 10μM
|
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
1400 |
0.55 |
Binding ≤ 10μM
|
|
BLAC-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
6500 |
0.48 |
Binding ≤ 10μM
|
|
BLAT-3-B |
Beta-lactamase TEM (cluster #3 Of 4), Bacterial |
Bacteria |
800 |
0.57 |
Binding ≤ 10μM
|
|
D1MIX9-1-B |
Beta-lactamase GES-13 (cluster #1 Of 1), Bacterial |
Bacteria |
370 |
0.60 |
Binding ≤ 10μM
|
|
D5HSX5-1-B |
Extended-spectrum Beta-lactamase PER-6 (cluster #1 Of 1), Bacterial |
Bacteria |
4000 |
0.50 |
Binding ≤ 10μM
|
|
Q0VTR6-1-B |
ADC-14 Protein (cluster #1 Of 1), Bacterial |
Bacteria |
1120 |
0.56 |
Binding ≤ 10μM
|
|
Q0VTR8-1-B |
ADC-16 Protein (cluster #1 Of 1), Bacterial |
Bacteria |
7758 |
0.48 |
Binding ≤ 10μM
|
|
Q7X575-1-B |
Beta-lactamase SHV-11 (cluster #1 Of 1), Bacterial |
Bacteria |
2400 |
0.52 |
Binding ≤ 10μM
|
|
Z50185-1-O |
Staphylococcus Aureus (cluster #1 Of 1), Other |
Other |
6800 |
0.48 |
Binding ≤ 10μM |
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 1), Other |
Other |
1900 |
0.53 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-0.6 |
-51.76 |
0 |
6 |
-1 |
95 |
232.237 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.65 |
6.84 |
-38.29 |
3 |
3 |
1 |
46 |
292.443 |
7 |
↓
|
|
|
|
Analogs
-
8214681
-
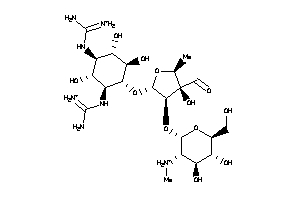
-
8214760
-
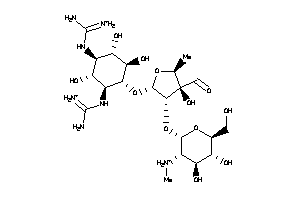
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TARB1-1-E |
TAR RNA Binding Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9500 |
0.18 |
Binding ≤ 10μM |
|
TRBP2-1-E |
TAR RNA Binding Protein 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9500 |
0.18 |
Binding ≤ 10μM |
|
Z50212-7-O |
Escherichia Coli (cluster #7 Of 7), Other |
Other |
1050 |
0.21 |
Functional ≤ 10μM
|
|
Z50380-2-O |
Mycobacterium Smegmatis (cluster #2 Of 4), Other |
Other |
500 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.28 |
-15.28 |
-142.69 |
19 |
19 |
3 |
339 |
584.604 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.28 |
-16.72 |
-90.55 |
18 |
19 |
2 |
335 |
583.596 |
11 |
↓
|
|
|
|
Analogs
-
8214392
-
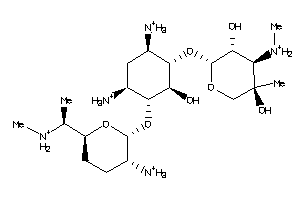
-
38139392
-
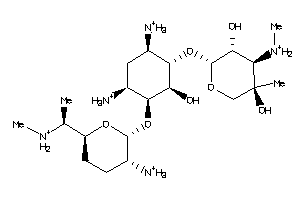
-
38139393
-
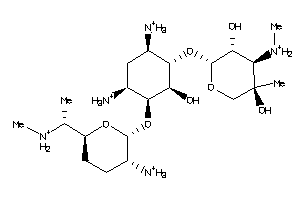
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-7-O |
Escherichia Coli (cluster #7 Of 7), Other |
Other |
30 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.21 |
-7.42 |
-414.09 |
16 |
12 |
5 |
214 |
482.643 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.21 |
-9.07 |
-99.13 |
13 |
12 |
2 |
206 |
479.619 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.21 |
-8.66 |
-180.42 |
14 |
12 |
3 |
208 |
480.627 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-2-O |
Escherichia Coli (cluster #2 Of 7), Other |
Other |
9000 |
0.64 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-0.54 |
-355.39 |
0 |
6 |
-4 |
126 |
240.859 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.97 |
-1.63 |
-218.59 |
1 |
6 |
-3 |
124 |
241.867 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.97 |
-2.73 |
-107.25 |
2 |
6 |
-2 |
121 |
242.875 |
2 |
↓
|
|
|
|
Analogs
-
608059
-
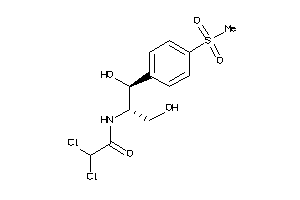
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
400 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.36 |
-7.65 |
-16.88 |
3 |
6 |
0 |
103 |
356.227 |
6 |
↓
|
|
|
|
Analogs
-
1622
-
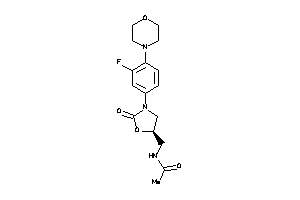
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
950 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
4.03 |
-20.29 |
1 |
7 |
0 |
71 |
337.351 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.73 |
2.77 |
-13.25 |
3 |
7 |
0 |
115 |
323.132 |
6 |
↓
|
|
|
|
Analogs
-
16248873
-
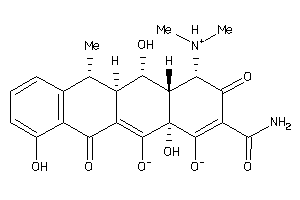
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.43 |
0.98 |
-89.55 |
6 |
10 |
-1 |
188 |
443.432 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-0.56 |
-4.34 |
-53.29 |
6 |
10 |
-1 |
185 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.61 |
-2.24 |
-69.99 |
6 |
10 |
-1 |
184 |
443.432 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
0.59 |
-95.46 |
6 |
10 |
-1 |
188 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
-1.58 |
-146.85 |
5 |
10 |
-2 |
187 |
442.424 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.24 |
-0.62 |
-247.57 |
4 |
10 |
-3 |
190 |
441.416 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.22 |
2.61 |
-130.09 |
3 |
15 |
-2 |
221 |
552.576 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100263-1-O |
Ribosomal RNA A-site (cluster #1 Of 1), Other |
Other |
1400 |
0.25 |
Binding ≤ 10μM
|
|
Z50212-7-O |
Escherichia Coli (cluster #7 Of 7), Other |
Other |
400 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.82 |
-21.35 |
-307.83 |
20 |
15 |
4 |
295 |
487.551 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.82 |
-22.1 |
-130.72 |
18 |
15 |
2 |
292 |
485.535 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.82 |
-22.05 |
-114.94 |
18 |
15 |
2 |
292 |
485.535 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-2-O |
Escherichia Coli (cluster #2 Of 7), Other |
Other |
510 |
0.98 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.21 |
-3.02 |
-228.45 |
1 |
6 |
-3 |
124 |
172.977 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.21 |
-1.93 |
-368.21 |
0 |
6 |
-4 |
126 |
171.969 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.21 |
-4.12 |
-114.07 |
2 |
6 |
-2 |
121 |
173.985 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DXR-2-B |
1-deoxyxylulose-5-phosphate Reductoisomerase (cluster #2 Of 2), Bacterial |
Bacteria |
21 |
0.98 |
Binding ≤ 10μM
|
|
Z50212-4-O |
Escherichia Coli (cluster #4 Of 7), Other |
Other |
220 |
0.85 |
Functional ≤ 10μM
|
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
480 |
0.80 |
Functional ≤ 10μM
|
|
Z50426-2-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #2 Of 9), Other |
Other |
3700 |
0.69 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.58 |
-1.63 |
-45.27 |
2 |
6 |
-1 |
101 |
182.092 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
3900 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
-1.27 |
-18.71 |
1 |
6 |
0 |
75 |
316.357 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-4-O |
Escherichia Coli (cluster #4 Of 7), Other |
Other |
580 |
0.79 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.44 |
-2.59 |
-194.69 |
2 |
7 |
-3 |
143 |
202.019 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.44 |
-3.7 |
-111.02 |
3 |
7 |
-2 |
140 |
203.027 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.44 |
-4.82 |
-44.14 |
4 |
7 |
-1 |
137 |
204.035 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-5-O |
Escherichia Coli (cluster #5 Of 7), Other |
Other |
68 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.62 |
4.73 |
-18.17 |
0 |
6 |
0 |
71 |
294.18 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-6-O |
Escherichia Coli (cluster #6 Of 7), Other |
Other |
9500 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.27 |
-4.05 |
-16.22 |
2 |
6 |
0 |
84 |
204.157 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
35 |
0.33 |
Binding ≤ 10μM
|
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
320 |
0.28 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
4 |
0.37 |
Functional ≤ 10μM
|
|
Z80119-1-O |
Detroit 98 (cluster #1 Of 1), Other |
Other |
2 |
0.38 |
Functional ≤ 10μM
|
|
Z80171-1-O |
HuTu80 (cluster #1 Of 1), Other |
Other |
5 |
0.36 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7900 |
0.22 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
7 |
0.36 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
4 |
0.37 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2050 |
0.25 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
320 |
0.28 |
Functional ≤ 10μM
|
|
Z81131-1-O |
L Cells (Fibroblasts) (cluster #1 Of 1), Other |
Other |
2 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.21 |
4.48 |
-122.21 |
6 |
13 |
-2 |
225 |
438.404 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.21 |
4.93 |
-127.62 |
7 |
13 |
-1 |
226 |
439.412 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-6-O |
Escherichia Coli (cluster #6 Of 7), Other |
Other |
20 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.01 |
-8.86 |
-22.27 |
4 |
8 |
0 |
132 |
361.445 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-3-O |
Escherichia Coli (cluster #3 Of 7), Other |
Other |
5600 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
1.07 |
-10.42 |
2 |
3 |
0 |
53 |
207.254 |
1 |
↓
|
|
|
|
Analogs
-
4554508
-
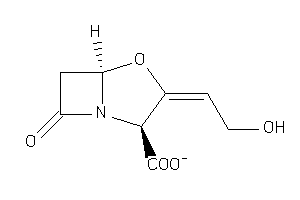
-
4554509
-
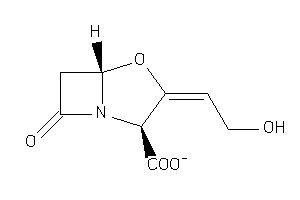
-
4554510
-
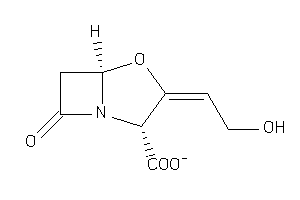
-
4554511
-
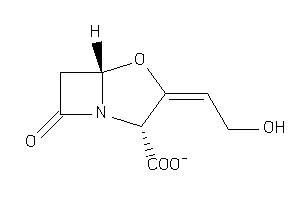
-
1169
-
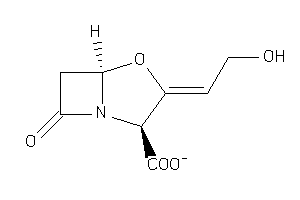
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A0ZX81-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
1720 |
0.58 |
Binding ≤ 10μM
|
|
A1E3K9-1-B |
Beta-lactamase SCO-1 (cluster #1 Of 1), Bacterial |
Bacteria |
70 |
0.72 |
Binding ≤ 10μM
|
|
A4KCT8-1-B |
Gil1 (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.80 |
Binding ≤ 10μM
|
|
A8RR46-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
85 |
0.71 |
Binding ≤ 10μM
|
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
800 |
0.61 |
Binding ≤ 10μM
|
|
BLA1-1-B |
Beta-lactamase L1 (cluster #1 Of 3), Bacterial |
Bacteria |
170 |
0.68 |
Binding ≤ 10μM
|
|
BLAC-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
80 |
0.71 |
Binding ≤ 10μM
|
|
BLAT-3-B |
Beta-lactamase TEM (cluster #3 Of 4), Bacterial |
Bacteria |
800 |
0.61 |
Binding ≤ 10μM
|
|
BLO1-1-B |
Beta-lactamase OXA-1 (cluster #1 Of 1), Bacterial |
Bacteria |
3200 |
0.55 |
Binding ≤ 10μM
|
|
D1MIX9-1-B |
Beta-lactamase GES-13 (cluster #1 Of 1), Bacterial |
Bacteria |
100 |
0.70 |
Binding ≤ 10μM
|
|
Q2XPY6-1-B |
Extended-spectrum Beta-lactamase CTX-M-53 (cluster #1 Of 1), Bacterial |
Bacteria |
10 |
0.80 |
Binding ≤ 10μM
|
|
Q4TVR4-1-B |
Beta-lactamase SHV-55 (cluster #1 Of 1), Bacterial |
Bacteria |
20 |
0.77 |
Binding ≤ 10μM
|
|
Q50H31-1-B |
Class D Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
2000 |
0.57 |
Binding ≤ 10μM
|
|
Q6GWS8-1-B |
Beta-lactamase TEM-125 (cluster #1 Of 1), Bacterial |
Bacteria |
8600 |
0.51 |
Binding ≤ 10μM
|
|
Q9EXV5-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.80 |
Binding ≤ 10μM
|
|
BLAT-1-B |
Beta-lactamase TEM (cluster #1 Of 1), Bacterial |
Bacteria |
60 |
0.72 |
Functional ≤ 10μM
|
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
40 |
0.74 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.42 |
-3.38 |
-49.41 |
1 |
6 |
-1 |
90 |
198.154 |
2 |
↓
|
|
|
|
Analogs
-
597112
-
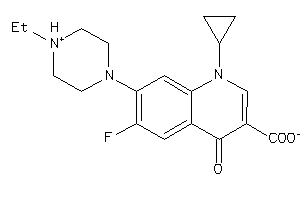
-
6385314
-
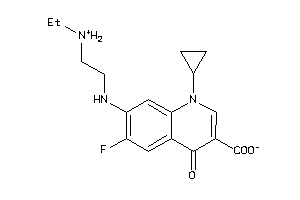
-
39294843
-
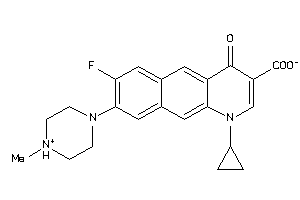
Draw
Identity
99%
90%
80%
70%
Vendors
And 85 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GYRA-1-B |
DNA Gyrase Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.37 |
Binding ≤ 10μM
|
|
GYRB-1-B |
DNA Gyrase Subunit B (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.37 |
Binding ≤ 10μM
|
|
PARC-1-B |
Topoisomerase IV Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
2300 |
0.33 |
Binding ≤ 10μM
|
|
GYRA-1-B |
DNA Gyrase Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
200 |
0.39 |
Functional ≤ 10μM
|
|
GYRB-1-B |
DNA Gyrase Subunit B (cluster #1 Of 1), Bacterial |
Bacteria |
200 |
0.39 |
Functional ≤ 10μM
|
|
PARC-1-B |
Topoisomerase IV Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
2500 |
0.33 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3020 |
0.32 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.44 |
Binding ≤ 10μM |
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.44 |
Binding ≤ 10μM |
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
410 |
0.37 |
Binding ≤ 10μM
|
|
Z50117-1-O |
Pseudomonas Aeruginosa (cluster #1 Of 2), Other |
Other |
220 |
0.39 |
Functional ≤ 10μM
|
|
Z50185-3-O |
Staphylococcus Aureus (cluster #3 Of 4), Other |
Other |
400 |
0.37 |
Functional ≤ 10μM
|
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
10 |
0.47 |
Functional ≤ 10μM
|
|
Z50378-1-O |
Mycobacterium Intracellulare (cluster #1 Of 1), Other |
Other |
910 |
0.35 |
Functional ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
1980 |
0.33 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
9440 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.70 |
9.07 |
-109.14 |
2 |
6 |
0 |
82 |
331.347 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.70 |
7.77 |
-65.4 |
1 |
6 |
-1 |
77 |
330.339 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.45 |
7.06 |
-88.49 |
3 |
6 |
1 |
85 |
332.355 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 43 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBR1-2-E |
Carbonyl Reductase [NADPH] 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.59 |
Binding ≤ 10μM
|
|
Q6UCJ9-1-E |
Enoyl-acyl Carrier Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.63 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.67 |
Binding ≤ 10μM
|
|
Q9BJJ9-3-E |
Enoyl-ACP Reductase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.55 |
Binding ≤ 10μM |
|
FABI-2-B |
Enoyl-[acyl-carrier-protein] Reductase (cluster #2 Of 2), Bacterial |
Bacteria |
70 |
0.59 |
Binding ≤ 10μM
|
|
INHA-4-B |
Enoyl-[acyl-carrier-protein] Reductase (cluster #4 Of 4), Bacterial |
Bacteria |
3000 |
0.45 |
Binding ≤ 10μM
|
|
Z50212-3-O |
Escherichia Coli (cluster #3 Of 7), Other |
Other |
750 |
0.50 |
Functional ≤ 10μM
|
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
774 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
6.75 |
-4 |
1 |
2 |
0 |
29 |
289.545 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.13 |
7.67 |
-38.31 |
0 |
2 |
-1 |
32 |
288.537 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50212-2-O |
Escherichia Coli (cluster #2 Of 7), Other |
Other |
330 |
0.82 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.43 |
-5.34 |
-225.71 |
2 |
7 |
-3 |
144 |
203.003 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.43 |
-4.24 |
-364.14 |
1 |
7 |
-4 |
147 |
201.995 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.43 |
-6.47 |
-111.47 |
3 |
7 |
-2 |
141 |
204.011 |
2 |
↓
|
|