|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
60 |
0.72 |
Binding ≤ 10μM
|
|
5HT1B-2-E |
Serotonin 1b (5-HT1b) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
-1.87 |
-45.39 |
3 |
2 |
1 |
36 |
188.25 |
3 |
↓
|
|
|
|
Analogs
-
34554993
-
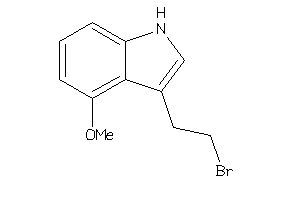
-
38266522
-
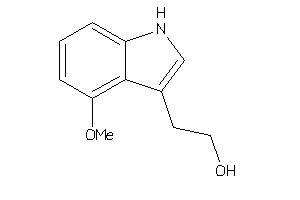
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT5B-1-E |
Serotonin 5b (5-HT5b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.08 |
-2.66 |
-44.39 |
4 |
3 |
1 |
52 |
191.254 |
3 |
↓
|
|
|
|
Analogs
-
3651042
-
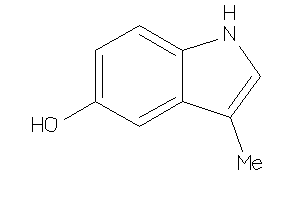
Draw
Identity
99%
90%
80%
70%
Vendors
And 69 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.89 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM
|
|
5HT1E-1-E |
Serotonin 1e (5-HT1e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.90 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
680 |
0.66 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
680 |
0.66 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
680 |
0.66 |
Binding ≤ 10μM
|
|
5HT3A-4-E |
Serotonin 3a (5-HT3a) Receptor (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
120 |
0.75 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.76 |
Binding ≤ 10μM |
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.75 |
Binding ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.75 |
Binding ≤ 10μM
|
|
5HT3E-1-E |
Serotonin 3e (5-HT3e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.75 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.80 |
Binding ≤ 10μM |
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.71 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
93 |
0.76 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.87 |
Binding ≤ 10μM
|
|
AA3R-4-E |
Adenosine Receptor A3 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
79 |
0.76 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9690 |
0.54 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
523 |
0.68 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
740 |
0.66 |
Binding ≤ 10μM
|
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.86 |
Functional ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.89 |
Functional ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.89 |
Functional ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.87 |
Functional ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.77 |
Functional ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Functional ≤ 10μM
|
|
5HT2C-1-E |
5-hydroxytryptamine Receptor 2C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Functional ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.68 |
Functional ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.90 |
Functional ≤ 10μM |
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.84 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50588-4-O |
Canis Familiaris (cluster #4 Of 7), Other |
Other |
210 |
0.72 |
Functional ≤ 10μM
|
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 8), Other |
Other |
300 |
0.70 |
Functional ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
919 |
0.65 |
Functional ≤ 10μM
|
|
Z80334-1-O |
NG108-15 (Neuroblastoma-glioma Hybrid Cells) (cluster #1 Of 1), Other |
Other |
298 |
0.70 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
0.5 |
-47.88 |
5 |
3 |
1 |
64 |
177.227 |
2 |
↓
|
|
|
|
Analogs
-
1530638
-
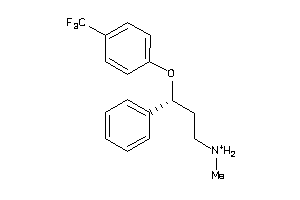
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
130 |
0.44 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5820 |
0.33 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
176 |
0.43 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
563 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.51 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8300 |
0.32 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
605 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.34 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
7000 |
0.33 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
33 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
8 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
9.16 |
-43.81 |
2 |
2 |
1 |
26 |
310.339 |
7 |
↓
|
|
|
|
Analogs
-
1530637
-
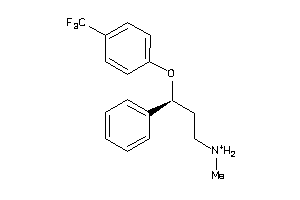
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
130 |
0.44 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5820 |
0.33 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
176 |
0.43 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
563 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.51 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
31 |
0.48 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8300 |
0.32 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
605 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.34 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
7000 |
0.33 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
21 |
0.49 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
8 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
9.16 |
-44.31 |
2 |
2 |
1 |
26 |
310.339 |
7 |
↓
|
|
|
|
Analogs
-
2988838
-
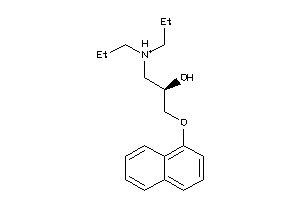
-
32111148
-
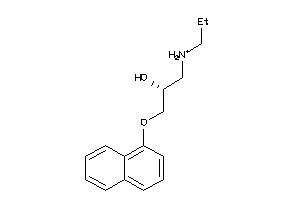
-
32111149
-
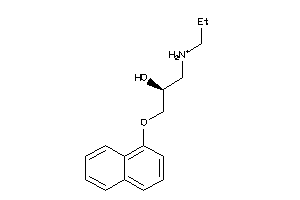
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1325 |
0.37 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
0.24 |
-39.91 |
2 |
3 |
1 |
33 |
302.438 |
9 |
↓
|
|
|
|
Analogs
-
2988837
-
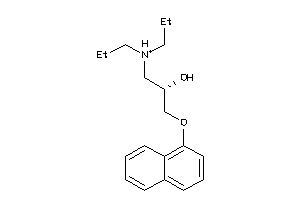
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1325 |
0.37 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
9.15 |
-39.83 |
2 |
3 |
1 |
34 |
302.438 |
9 |
↓
|
|
|
|
Analogs
-
4071759
-
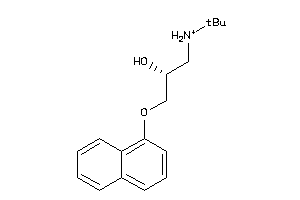
-
32102194
-
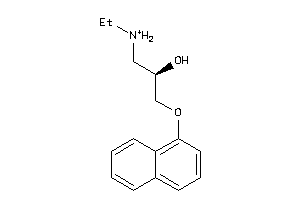
-
32102197
-
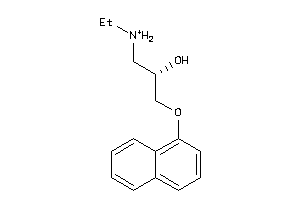
-
20240
-
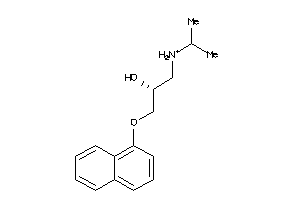
Draw
Identity
99%
90%
80%
70%
Vendors
And 69 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
55 |
0.53 |
Binding ≤ 10μM
|
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.52 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.48 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.48 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8100 |
0.38 |
Binding ≤ 10μM
|
|
5HT2B-3-E |
Serotonin 2b (5-HT2b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.39 |
Binding ≤ 10μM
|
|
5HT2C-3-E |
Serotonin 2c (5-HT2c) Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.43 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9700 |
0.37 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
5HT1B-2-E |
Serotonin 1b (5-HT1b) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.40 |
Functional ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.43 |
ADME/T ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.42 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.48 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.47 |
ADME/T ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
3162 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.47 |
-42.1 |
3 |
3 |
1 |
46 |
260.357 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.97 |
4.28 |
-6.82 |
2 |
3 |
0 |
41 |
259.349 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
3.19 |
-42.74 |
4 |
4 |
1 |
62 |
249.334 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.98 |
2.01 |
-8.94 |
3 |
4 |
0 |
57 |
248.326 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
3.18 |
-42.7 |
4 |
4 |
1 |
62 |
249.334 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.98 |
1.9 |
-9.07 |
3 |
4 |
0 |
57 |
248.326 |
6 |
↓
|
|
|
|
Analogs
-
19843876
-
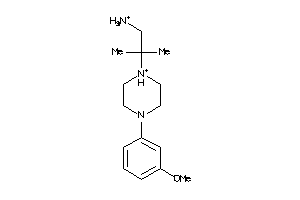
-
34989992
-
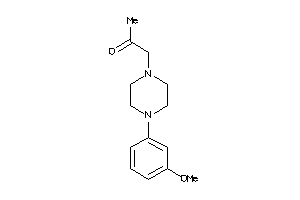
Draw
Identity
99%
90%
80%
70%
Vendors
And 75 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
2.66 |
-43 |
2 |
3 |
1 |
29 |
193.27 |
2 |
↓
|
|
|
|
Analogs
-
34670106
-
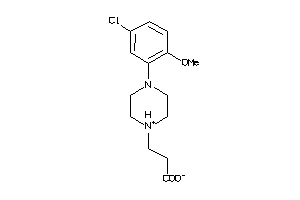
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
870 |
0.57 |
Binding ≤ 10μM
|
|
5HT3A-2-E |
Serotonin 3a (5-HT3a) Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
40 |
0.69 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
40 |
0.69 |
Binding ≤ 10μM
|
|
5HT3C-2-E |
Serotonin 3c (5-HT3c) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.69 |
Binding ≤ 10μM
|
|
5HT3D-2-E |
Serotonin 3d (5-HT3d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.69 |
Binding ≤ 10μM
|
|
5HT3E-2-E |
Serotonin 3e (5-HT3e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
3.45 |
-41.46 |
2 |
3 |
1 |
29 |
227.715 |
2 |
↓
|
|
|
|
Analogs
-
38256242
-
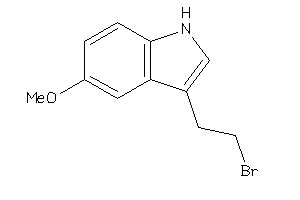
-
39234508
-
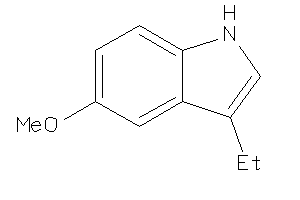
Draw
Identity
99%
90%
80%
70%
Vendors
And 108 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.80 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.64 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.83 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
690 |
0.62 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.73 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.76 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
88 |
0.71 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
2.65 |
-46.13 |
4 |
3 |
1 |
53 |
191.254 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
2580783
-
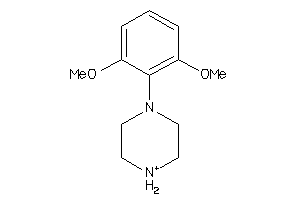
-
34434918
-
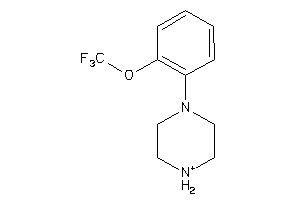
-
36472586
-
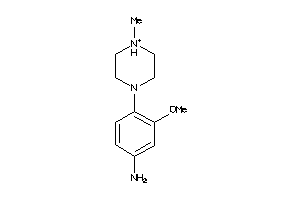
Draw
Identity
99%
90%
80%
70%
Vendors
And 105 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.80 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.75 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.75 |
Binding ≤ 10μM
|
|
5HT1E-2-E |
Serotonin 1e (5-HT1e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.75 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.75 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.55 |
Binding ≤ 10μM
|
|
5HT2B-3-E |
Serotonin 2b (5-HT2b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3500 |
0.55 |
Binding ≤ 10μM
|
|
5HT2C-3-E |
Serotonin 2c (5-HT2c) Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.55 |
Binding ≤ 10μM
|
|
5HT3A-2-E |
Serotonin 3a (5-HT3a) Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
5HT3C-2-E |
Serotonin 3c (5-HT3c) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
5HT3D-2-E |
Serotonin 3d (5-HT3d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
5HT3E-2-E |
Serotonin 3e (5-HT3e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.90 |
Binding ≤ 10μM
|
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3508 |
0.55 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.57 |
Binding ≤ 10μM
|
|
DRD2-11-E |
Dopamine D2 Receptor (cluster #11 Of 24), Eukaryotic |
Eukaryotes |
1200 |
0.59 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
510 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
4.29 |
-39.16 |
2 |
3 |
1 |
29 |
193.27 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.35 |
2.98 |
-3.93 |
1 |
3 |
0 |
24 |
192.262 |
2 |
↓
|
|
|
|
Analogs
-
32113702
-
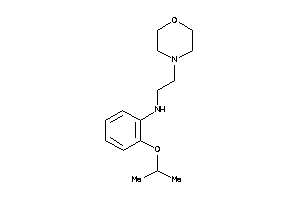
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
4.53 |
-41.98 |
2 |
3 |
1 |
29 |
221.324 |
3 |
↓
|
|
|
|
Analogs
-
7653965
-
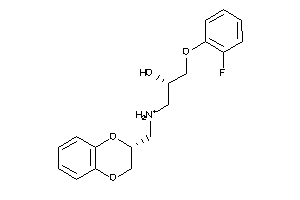
-
7653967
-
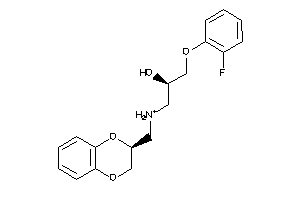
-
7653971
-
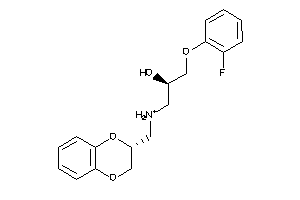
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
197 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
5.03 |
-54.31 |
3 |
5 |
1 |
65 |
334.367 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.04 |
3.66 |
-14.22 |
2 |
5 |
0 |
60 |
333.359 |
7 |
↓
|
|
|
|
Analogs
-
7653967
-

-
7653971
-

-
7653962
-
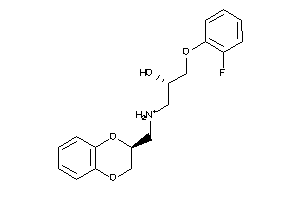
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
197 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
5 |
-54.46 |
3 |
5 |
1 |
65 |
334.367 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.04 |
3.66 |
-13.36 |
2 |
5 |
0 |
60 |
333.359 |
7 |
↓
|
|
|
|
Analogs
-
7653971
-

-
7653965
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
197 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
5 |
-54.48 |
3 |
5 |
1 |
65 |
334.367 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.04 |
3.55 |
-14.23 |
2 |
5 |
0 |
60 |
333.359 |
7 |
↓
|
|
|
|
Analogs
-
7653965
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
197 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
4.98 |
-54.43 |
3 |
5 |
1 |
65 |
334.367 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.04 |
3.55 |
-13.48 |
2 |
5 |
0 |
60 |
333.359 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
36 |
0.42 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
265 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
-3.83 |
-48.75 |
3 |
5 |
1 |
64 |
364.849 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
36 |
0.42 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
265 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
-3.78 |
-48.58 |
3 |
5 |
1 |
64 |
364.849 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
36 |
0.42 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
265 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
6.17 |
-52.96 |
3 |
5 |
1 |
65 |
364.849 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.46 |
4.72 |
-10.67 |
2 |
5 |
0 |
60 |
363.841 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
36 |
0.42 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
265 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
6.22 |
-53.2 |
3 |
5 |
1 |
65 |
364.849 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.46 |
4.66 |
-10.08 |
2 |
5 |
0 |
60 |
363.841 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
88 |
0.62 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.73 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
4.38 |
-45.99 |
4 |
3 |
1 |
53 |
219.308 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
24 |
0.36 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.39 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
348 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
6.59 |
-12.25 |
1 |
6 |
0 |
62 |
425.554 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
2169424
-
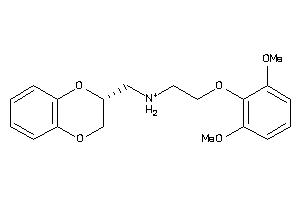
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
46 |
0.41 |
Binding ≤ 10μM
|
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
107 |
0.39 |
Binding ≤ 10μM
|
|
DRD2-3-E |
Dopamine D2 Receptor (cluster #3 Of 24), Eukaryotic |
Eukaryotes |
123 |
0.39 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
9 |
0.45 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
5012 |
0.30 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
7762 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
7.08 |
-42.51 |
2 |
6 |
1 |
63 |
346.403 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.20 |
5.74 |
-11.86 |
1 |
6 |
0 |
58 |
345.395 |
8 |
↓
|
|
|
|
Analogs
-
2169422
-
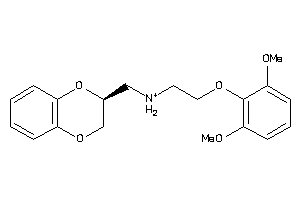
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
46 |
0.41 |
Binding ≤ 10μM
|
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
107 |
0.39 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
9 |
0.45 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
5012 |
0.30 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
7762 |
0.29 |
Functional ≤ 10μM
|
|
DRD2-3-E |
Dopamine D2 Receptor (cluster #3 Of 24), Eukaryotic |
Eukaryotes |
123 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
7.06 |
-41.62 |
2 |
6 |
1 |
63 |
346.403 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.20 |
5.67 |
-11.58 |
1 |
6 |
0 |
58 |
345.395 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.73 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.73 |
Binding ≤ 10μM
|
|
5HT1D-2-E |
Serotonin 1d (5-HT1d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.73 |
Binding ≤ 10μM
|
|
5HT1E-2-E |
Serotonin 1e (5-HT1e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.73 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.73 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
62 |
0.63 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
62 |
0.63 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
62 |
0.63 |
Binding ≤ 10μM
|
|
5HT3A-2-E |
Serotonin 3a (5-HT3a) Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
175 |
0.59 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
175 |
0.59 |
Binding ≤ 10μM
|
|
5HT3C-2-E |
Serotonin 3c (5-HT3c) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
175 |
0.59 |
Binding ≤ 10μM
|
|
5HT3D-2-E |
Serotonin 3d (5-HT3d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
175 |
0.59 |
Binding ≤ 10μM
|
|
5HT3E-2-E |
Serotonin 3e (5-HT3e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
175 |
0.59 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.65 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.61 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
83 |
0.62 |
Binding ≤ 10μM
|
|
SGMR1-4-E |
Sigma Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
2720 |
0.49 |
Binding ≤ 10μM
|
|
Z50425-7-O |
Plasmodium Falciparum (cluster #7 Of 22), Other |
Other |
7943 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
7.56 |
-41.85 |
2 |
2 |
1 |
20 |
213.304 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.50 |
6.06 |
-3.52 |
1 |
2 |
0 |
15 |
212.296 |
1 |
↓
|
|
|
|
Analogs
-
6274060
-
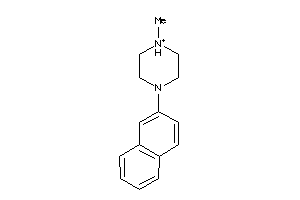
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2900 |
0.48 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
265 |
0.58 |
Binding ≤ 10μM
|
|
5HT1D-2-E |
Serotonin 1d (5-HT1d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
265 |
0.58 |
Binding ≤ 10μM
|
|
5HT1E-2-E |
Serotonin 1e (5-HT1e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
265 |
0.58 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
265 |
0.58 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.63 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.63 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.63 |
Binding ≤ 10μM
|
|
5HT3A-2-E |
Serotonin 3a (5-HT3a) Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
32 |
0.66 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
32 |
0.66 |
Binding ≤ 10μM
|
|
5HT3C-2-E |
Serotonin 3c (5-HT3c) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
32 |
0.66 |
Binding ≤ 10μM
|
|
5HT3D-2-E |
Serotonin 3d (5-HT3d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
32 |
0.66 |
Binding ≤ 10μM
|
|
5HT3E-2-E |
Serotonin 3e (5-HT3e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
32 |
0.66 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.50 |
Binding ≤ 10μM
|
|
SGMR1-4-E |
Sigma Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
550 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
-2.87 |
-43.55 |
2 |
2 |
1 |
20 |
213.304 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.48 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.45 |
Binding ≤ 10μM
|
|
TRPM8-1-E |
Transient Receptor Potential Cation Channel Subfamily M Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5960 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
1.39 |
-38.2 |
2 |
3 |
1 |
29 |
295.406 |
6 |
↓
|
|
|
|
Analogs
-
13679044
-
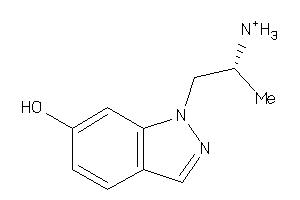
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
141 |
0.69 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7870 |
0.51 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.79 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.81 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.85 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
515 |
0.63 |
Binding ≤ 10μM
|
|
ADA2A-2-E |
Alpha-2a Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2270 |
0.56 |
Binding ≤ 10μM
|
|
ADA2C-2-E |
Alpha-2c Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4700 |
0.53 |
Binding ≤ 10μM
|
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1460 |
0.58 |
Functional ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
43 |
0.74 |
Functional ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Functional ≤ 10μM
|
|
5HT2C-1-E |
5-hydroxytryptamine Receptor 2C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Functional ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.53 |
Functional ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.56 |
Functional ≤ 10μM
|
|
ADA2B-2-E |
Alpha-2b Adrenergic Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3050 |
0.55 |
Functional ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.40 |
-3.42 |
-39.34 |
4 |
4 |
1 |
65 |
192.242 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
316 |
0.29 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.30 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
316 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
11.38 |
-40.48 |
1 |
5 |
1 |
43 |
413.545 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.59 |
11.44 |
-44.74 |
1 |
5 |
1 |
43 |
413.545 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.59 |
9.07 |
-12.22 |
0 |
5 |
0 |
41 |
412.537 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
128 |
0.51 |
Binding ≤ 10μM
|
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
554 |
0.46 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4730 |
0.39 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1880 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
7.83 |
-40.31 |
2 |
3 |
1 |
29 |
255.341 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.04 |
6.52 |
-4.83 |
1 |
3 |
0 |
24 |
254.333 |
3 |
↓
|
|
|
|
Analogs
-
43461478
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
31 |
0.50 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
526 |
0.42 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
978 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
8.44 |
-42.89 |
2 |
3 |
1 |
29 |
287.358 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
|
DRD2-11-E |
Dopamine D2 Receptor (cluster #11 Of 24), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
10.6 |
-43.03 |
1 |
4 |
1 |
26 |
345.438 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.06 |
8.4 |
-6.58 |
0 |
4 |
0 |
25 |
344.43 |
7 |
↓
|
|
|
|
Analogs
-
5850128
-
-
33809290
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4850 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
1.88 |
-36.52 |
1 |
2 |
1 |
13 |
180.271 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3300 |
0.64 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.64 |
Binding ≤ 10μM
|
|
5HT1B-2-E |
Serotonin 1b (5-HT1b) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3450 |
0.64 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
4.78 |
-40.29 |
2 |
2 |
1 |
26 |
166.244 |
5 |
↓
|
|
|
|
Analogs
-
34962480
-
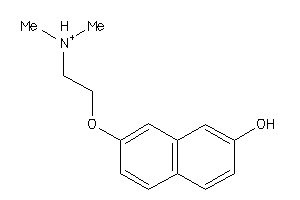
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.62 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.51 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.47 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.47 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.47 |
Binding ≤ 10μM
|
|
5HT1B-2-E |
Serotonin 1b (5-HT1b) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
94 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
2.12 |
-36.57 |
1 |
2 |
1 |
13 |
216.304 |
4 |
↓
|
|
|
|
Analogs
-
32123093
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
95 |
0.45 |
Binding ≤ 10μM
|
|
5HT1B-2-E |
Serotonin 1b (5-HT1b) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.42 |
Functional ≤ 10μM
|
|
5HT1D-2-E |
Serotonin 1d (5-HT1d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
590 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
12.08 |
-40.93 |
1 |
2 |
1 |
14 |
292.402 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.58 |
9.7 |
-6.36 |
0 |
2 |
0 |
12 |
291.394 |
6 |
↓
|
|
|
|
Analogs
-
36472586
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1035 |
0.52 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1035 |
0.52 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1035 |
0.52 |
Binding ≤ 10μM
|
|
5HT1E-2-E |
Serotonin 1e (5-HT1e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1035 |
0.52 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1035 |
0.52 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8430 |
0.44 |
Binding ≤ 10μM
|
|
5HT2B-3-E |
Serotonin 2b (5-HT2b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8430 |
0.44 |
Binding ≤ 10μM
|
|
5HT2C-3-E |
Serotonin 2c (5-HT2c) Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8430 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.38 |
2.23 |
-41.62 |
2 |
4 |
1 |
38 |
223.296 |
3 |
↓
|
|
|
|
Analogs
-
33809296
-
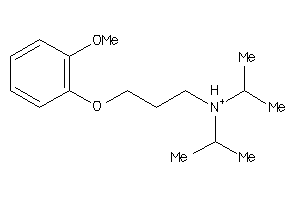
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
6.86 |
-38.52 |
1 |
3 |
1 |
23 |
210.297 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3420 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
2.28 |
-36.03 |
1 |
1 |
1 |
4 |
186.278 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
35 |
0.80 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
74 |
0.77 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
5HT1E-2-E |
Serotonin 1e (5-HT1e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
215 |
0.72 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
516 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
0.86 |
-42.83 |
3 |
3 |
1 |
40 |
179.243 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
56556
-
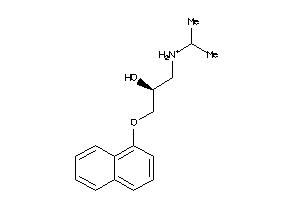
-
4071759
-

-
32102194
-

-
32102197
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 72 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.52 |
Binding ≤ 10μM
|
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.43 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9700 |
0.37 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.42 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.47 |
ADME/T ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
3162 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.45 |
-42.19 |
3 |
3 |
1 |
46 |
260.357 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.97 |
4.16 |
-6.9 |
2 |
3 |
0 |
41 |
259.349 |
6 |
↓
|
|