|
|
Analogs
-
9585
-
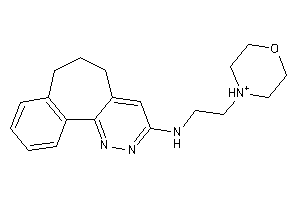
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
370 |
0.38 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
5.61 |
-10.41 |
1 |
5 |
0 |
50 |
324.428 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.02 |
8.05 |
-101.94 |
3 |
5 |
2 |
53 |
326.444 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
179 |
0.30 |
Binding ≤ 10μM |
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
73 |
0.32 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.24 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
9800 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
11.36 |
-42.06 |
2 |
7 |
1 |
75 |
422.553 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.13 |
8 |
-14.41 |
1 |
7 |
0 |
74 |
421.545 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.13 |
9.44 |
-87.43 |
3 |
7 |
2 |
77 |
423.561 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
179 |
0.30 |
Binding ≤ 10μM |
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
73 |
0.32 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.24 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
9800 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
9.44 |
-46.61 |
2 |
7 |
1 |
75 |
422.553 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.13 |
9.32 |
-14.39 |
1 |
7 |
0 |
74 |
421.545 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.13 |
9.54 |
-95.84 |
3 |
7 |
2 |
77 |
423.561 |
6 |
↓
|
|
|
|
Analogs
-
6582254
-
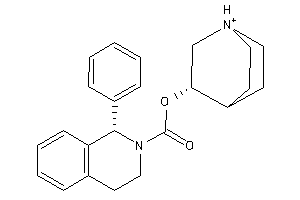
-
11616187
-
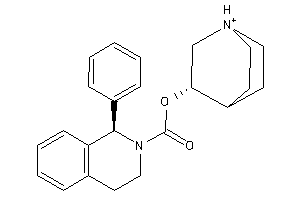
-
11616188
-
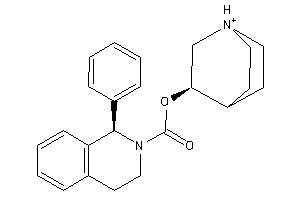
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
64 |
0.37 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
28 |
0.39 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
120 |
0.36 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.43 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
131 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
12.7 |
-38.95 |
1 |
4 |
1 |
34 |
363.481 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.51 |
8 |
-24.7 |
2 |
4 |
1 |
49 |
377.489 |
4 |
↓
|
|
|
|
Analogs
-
40164019
-
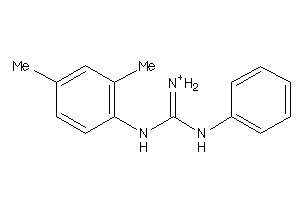
-
40164021
-
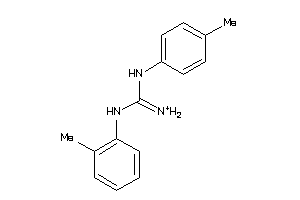
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
744 |
0.48 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2960 |
0.43 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7660 |
0.40 |
Binding ≤ 10μM
|
|
NMD3A-6-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMD3B-6-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE1-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-6-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDZ1-6-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
OPRD-3-E |
Delta Opioid Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
OPRK-2-E |
Kappa Opioid Receptor (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
SGMR1-4-E |
Sigma Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
89 |
0.55 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9210 |
0.39 |
Functional ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
2000 |
0.44 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
70 |
0.56 |
Binding ≤ 10μM
|
|
Z104302-7-O |
Glutamate NMDA Receptor (cluster #7 Of 7), Other |
Other |
9120 |
0.39 |
Binding ≤ 10μM
|
|
Z50512-3-O |
Cavia Porcellus (cluster #3 Of 7), Other |
Other |
6900 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
8.4 |
-25.53 |
4 |
3 |
1 |
50 |
240.33 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.71 |
10.09 |
-33.85 |
3 |
3 |
1 |
40 |
323.46 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5100 |
0.49 |
Binding ≤ 10μM
|
|
ADA2A-2-E |
Alpha-2a Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
950 |
0.56 |
Binding ≤ 10μM
|
|
ADA2B-2-E |
Alpha-2b Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
950 |
0.56 |
Binding ≤ 10μM
|
|
ADA2C-2-E |
Alpha-2c Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
950 |
0.56 |
Binding ≤ 10μM
|
|
Z104303-4-O |
Muscarinic Acetylcholine Receptor (cluster #4 Of 7), Other |
Other |
8800 |
0.47 |
Binding ≤ 10μM
|
|
Z104304-3-O |
Adrenergic Receptor Alpha-1 (cluster #3 Of 3), Other |
Other |
5900 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.24 |
7.73 |
-5.88 |
0 |
3 |
0 |
21 |
199.257 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.24 |
8.01 |
-33.75 |
1 |
3 |
1 |
22 |
200.265 |
3 |
↓
|
|
|
|
Analogs
-
32131087
-
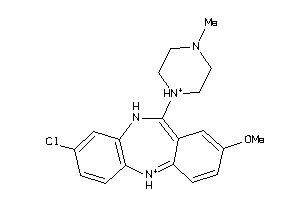
-
32131089
-
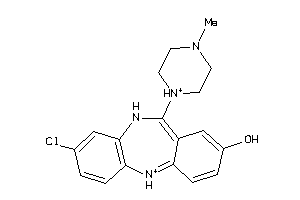
-
3995781
-
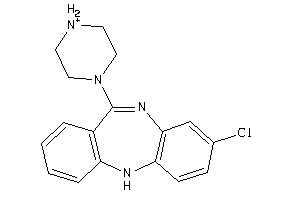
-
4292831
-
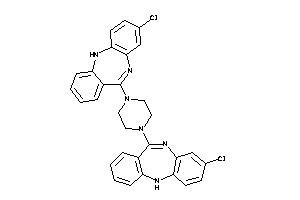
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.35 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT1D-2-E |
Serotonin 1d (5-HT1d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.49 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
156 |
0.41 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
34 |
0.45 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
34 |
0.45 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.31 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
960 |
0.37 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
890 |
0.37 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH4-1-E |
Histamine H4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.44 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6490 |
0.32 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
390 |
0.39 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
697 |
0.37 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3900 |
0.33 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8500 |
0.31 |
Binding ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
631 |
0.38 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
191 |
0.41 |
Functional ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
51 |
0.44 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
9 |
0.49 |
Binding ≤ 10μM
|
|
Z102275-1-O |
Hypothalamus (cluster #1 Of 1), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.55 |
-7.08 |
1 |
4 |
0 |
35 |
326.831 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.14 |
8.88 |
-23.43 |
2 |
4 |
1 |
36 |
327.839 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.14 |
11.23 |
-89.99 |
3 |
4 |
2 |
38 |
328.847 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
34 |
0.31 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.23 |
14.09 |
-85.07 |
3 |
7 |
2 |
77 |
465.642 |
9 |
↓
|
|
|
|
Analogs
-
3936683
-
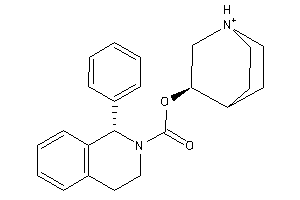
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
9 |
0.42 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
28 |
0.39 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
0.5 |
-37.73 |
1 |
4 |
1 |
34 |
363.481 |
3 |
↓
|
|
|
|
Analogs
-
75126
-
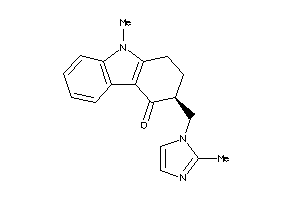
-
44699629
-
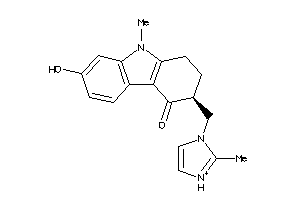
-
44699631
-
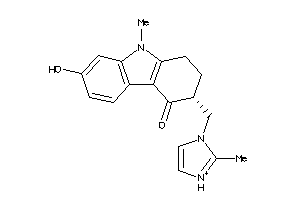
Draw
Identity
99%
90%
80%
70%
Vendors
And 69 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.35 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7800 |
0.33 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.33 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7800 |
0.33 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
5HT3E-1-E |
Serotonin 3e (5-HT3e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
960 |
0.38 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
910 |
0.38 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5400 |
0.34 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.34 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.39 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
810 |
0.39 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
813 |
0.39 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8500 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
680 |
0.39 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 2), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
240 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
11.15 |
-37.04 |
1 |
4 |
1 |
41 |
294.378 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.37 |
10.68 |
-21.62 |
0 |
4 |
0 |
40 |
293.37 |
2 |
↓
|
|
|
|
Analogs
-
44699629
-

-
44699631
-

-
4448
-
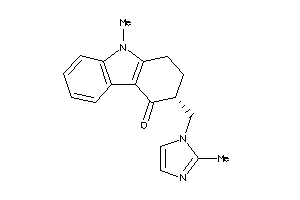
Draw
Identity
99%
90%
80%
70%
Vendors
And 73 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.35 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7800 |
0.33 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.33 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7800 |
0.33 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
5HT3E-1-E |
Serotonin 3e (5-HT3e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
960 |
0.38 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
910 |
0.38 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5400 |
0.34 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.34 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.39 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
813 |
0.39 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8500 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
680 |
0.39 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 2), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
240 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
11.1 |
-36.38 |
1 |
4 |
1 |
41 |
294.378 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.37 |
10.62 |
-21.13 |
0 |
4 |
0 |
40 |
293.37 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
720 |
0.54 |
Binding ≤ 10μM
|
|
NMD3A-6-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.71 |
Binding ≤ 10μM
|
|
NMD3B-6-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.71 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.71 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.71 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
5800 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
6.57 |
-34.7 |
1 |
2 |
1 |
17 |
211.288 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
5750285
-
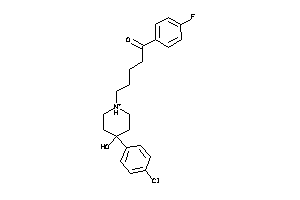
-
36464350
-
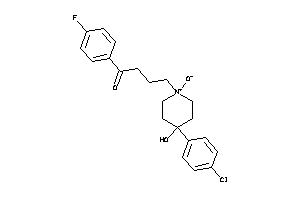
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.28 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.34 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.34 |
Binding ≤ 10μM
|
|
5HT1E-1-E |
Serotonin 1e (5-HT1e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.34 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.34 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.41 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
27 |
0.41 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
263 |
0.35 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5500 |
0.28 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6120 |
0.28 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4670 |
0.29 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4670 |
0.29 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4670 |
0.29 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
19 |
0.42 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9600 |
0.27 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9600 |
0.27 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9600 |
0.27 |
Binding ≤ 10μM
|
|
AP2S1-1-E |
AP-2 Complex Subunit Sigma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.41 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.34 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
DRD4-3-E |
Dopamine D4 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.43 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.36 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
730 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7520 |
0.28 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5300 |
0.28 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5687 |
0.28 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5687 |
0.28 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5687 |
0.28 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.28 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.31 |
Binding ≤ 10μM
|
|
SCN1A-2-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A-2-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.32 |
Binding ≤ 10μM
|
|
SCN8A-2-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
550 |
0.34 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
28 |
0.41 |
Functional ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
1 |
0.48 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
70 |
0.39 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
7466 |
0.28 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
9 |
0.43 |
Binding ≤ 10μM
|
|
Z103204-1-O |
A427 (cluster #1 Of 4), Other |
Other |
10000 |
0.27 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.30 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
7000 |
0.28 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM
|
|
Z80091-1-O |
CHO-hD3 (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Q4U254-1-V |
Human Rhinovirus A Protease (cluster #1 Of 3), Viral |
Viruses |
100 |
0.38 |
Binding ≤ 10μM
|
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
100 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
10.74 |
-49.71 |
2 |
3 |
1 |
42 |
376.879 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.48 |
Binding ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
12.49 |
-41.41 |
1 |
6 |
1 |
77 |
335.424 |
9 |
↓
|
|
|
|
Analogs
-
596
-
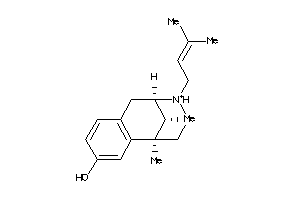
Draw
Identity
99%
90%
80%
70%
Popular Name:
(+-)-Pentazocine hydrochloride; (2R*,6R*,11R*)-1,2,3,4,5,6-Hexahydro-6,11-dimethyl-3-(3-methyl-2-butenyl)-2,6-methano-3-benzazocin-8-ol hydrochloride; (2alpha,6alpha,11R*)-1,2,3,4,5,6-Hexahydro-6,11-dimethyl-3-(3-methylbut-2-enyl)-2,6-methano-3-benzazocin
(+-)-Pentazocine hydrochloride; …
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
225 |
0.44 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
525 |
0.42 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
651 |
0.41 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.56 |
Binding ≤ 10μM |
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
651 |
0.41 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
860 |
0.40 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
4190 |
0.36 |
Binding ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
87 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
0.88 |
-37.21 |
2 |
2 |
1 |
24 |
286.439 |
2 |
↓
|
|
|
|
Analogs
-
11616187
-

-
11616188
-

-
3936683
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
230 |
0.34 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3500 |
0.28 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1300 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
0.91 |
-38.51 |
1 |
4 |
1 |
33 |
363.481 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.51 |
7.86 |
-26.12 |
2 |
4 |
1 |
49 |
377.489 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
610 |
0.40 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.51 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.51 |
Binding ≤ 10μM |
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
365 |
0.41 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM |
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM |
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM |
|
ACM4-3-E |
Muscarinic Acetylcholine Receptor M4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM |
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2870 |
0.35 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2870 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2870 |
0.35 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
52 |
0.46 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.46 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM |
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6740 |
0.33 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
958 |
0.38 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 24), Eukaryotic |
Eukaryotes |
69 |
0.46 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
88 |
0.45 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
10 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
6.89 |
-8.12 |
1 |
4 |
0 |
35 |
312.442 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
7240 |
0.38 |
Binding ≤ 10μM |
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5304 |
0.39 |
Binding ≤ 10μM |
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
2399 |
0.41 |
Binding ≤ 10μM
|
|
ACM4-4-E |
Muscarinic Acetylcholine Receptor M4 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
9120 |
0.37 |
Binding ≤ 10μM
|
|
ACM5-4-E |
Muscarinic Acetylcholine Receptor M5 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3802 |
0.40 |
Binding ≤ 10μM
|
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.40 |
Functional ≤ 10μM |
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6500 |
0.38 |
Functional ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
64 |
0.53 |
Binding ≤ 10μM |
|
Z50592-4-O |
Oryctolagus Cuniculus (cluster #4 Of 8), Other |
Other |
659 |
0.46 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.83 |
8.93 |
-38.89 |
1 |
4 |
1 |
38 |
281.763 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
6.24 |
-43.52 |
2 |
7 |
1 |
75 |
352.418 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.34 |
6.04 |
-38.61 |
2 |
7 |
1 |
75 |
352.418 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.34 |
3.86 |
-15.5 |
1 |
7 |
0 |
74 |
351.41 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
8900 |
0.26 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
8900 |
0.26 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8900 |
0.26 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8900 |
0.26 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8900 |
0.26 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
15 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
9.8 |
-53.2 |
2 |
4 |
1 |
51 |
363.481 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
8900 |
0.26 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
8900 |
0.26 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8900 |
0.26 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8900 |
0.26 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8900 |
0.26 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
15 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
9.76 |
-54.93 |
2 |
4 |
1 |
51 |
363.481 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1400 |
0.37 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5300 |
0.34 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.50 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1400 |
0.37 |
Binding ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6900 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
10.9 |
-42.69 |
1 |
2 |
1 |
17 |
311.474 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.05 |
8.42 |
-6.89 |
0 |
2 |
0 |
16 |
310.466 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.05 |
11.84 |
-92.52 |
2 |
2 |
2 |
19 |
312.482 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
19 |
0.49 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.51 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
2512 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.83 |
10.28 |
-36.84 |
2 |
2 |
1 |
25 |
302.482 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
19 |
0.49 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.51 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
2512 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.83 |
9.8 |
-38.56 |
2 |
2 |
1 |
25 |
302.482 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.51 |
Binding ≤ 10μM |
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
23 |
0.51 |
Binding ≤ 10μM |
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.51 |
Binding ≤ 10μM |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
333 |
0.43 |
Binding ≤ 10μM |
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
333 |
0.43 |
Binding ≤ 10μM |
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
333 |
0.43 |
Binding ≤ 10μM |
|
ACM4-3-E |
Muscarinic Acetylcholine Receptor M4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
333 |
0.43 |
Binding ≤ 10μM |
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.43 |
Binding ≤ 10μM |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.47 |
Binding ≤ 10μM |
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.47 |
Binding ≤ 10μM |
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.47 |
Binding ≤ 10μM |
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.40 |
Binding ≤ 10μM |
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.40 |
Binding ≤ 10μM |
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.40 |
Binding ≤ 10μM |
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
203 |
0.45 |
Binding ≤ 10μM |
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.51 |
Binding ≤ 10μM |
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 24), Eukaryotic |
Eukaryotes |
9 |
0.54 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
7.03 |
-49.22 |
3 |
4 |
1 |
49 |
299.423 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
2.88 |
8.06 |
-61.76 |
3 |
4 |
1 |
49 |
299.423 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.88 |
6.75 |
-10 |
2 |
4 |
0 |
44 |
298.415 |
1 |
↓
|
|
|
|
Analogs
-
597532
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.36 |
Binding ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
63 |
0.32 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9 |
0.35 |
Binding ≤ 10μM
|
|
ACM5-3-E |
Muscarinic Acetylcholine Receptor M5 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
11.71 |
-52.82 |
3 |
4 |
1 |
57 |
427.568 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.67 |
10.48 |
-13.04 |
2 |
4 |
0 |
56 |
426.56 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.49 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1200 |
0.49 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.49 |
Binding ≤ 10μM
|
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.70 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.70 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.70 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.70 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.70 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.70 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
-3.12 |
-30.1 |
3 |
3 |
1 |
43 |
226.303 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
870 |
0.47 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.47 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.47 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.47 |
Binding ≤ 10μM
|
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
6.08 |
-28.9 |
3 |
3 |
1 |
43 |
240.33 |
1 |
↓
|
|
|
|
Analogs
-
3929426
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
2900 |
0.22 |
Binding ≤ 10μM
|
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2340 |
0.23 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2340 |
0.23 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2340 |
0.23 |
Binding ≤ 10μM
|
|
CP2J2-1-E |
Cytochrome P450 2J2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8100 |
0.20 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
94 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
6700 |
0.21 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
563 |
0.25 |
Functional ≤ 10μM |
|
KCNH2-2-E |
HERG (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
204 |
0.27 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.23 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.26 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.21 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4950 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.17 |
14.49 |
-42.25 |
3 |
3 |
1 |
45 |
472.693 |
9 |
↓
|
|
|
|
Analogs
-
3929426
-

-
2383100
-
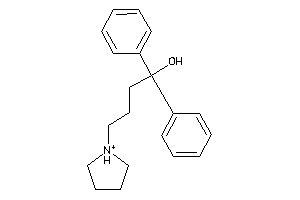
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
2900 |
0.22 |
Binding ≤ 10μM
|
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2340 |
0.23 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2340 |
0.23 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2340 |
0.23 |
Binding ≤ 10μM
|
|
CP2J2-1-E |
Cytochrome P450 2J2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8100 |
0.20 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
94 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
6700 |
0.21 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
563 |
0.25 |
Functional ≤ 10μM |
|
KCNH2-2-E |
HERG (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
204 |
0.27 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.23 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.26 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.21 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4950 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.17 |
14.46 |
-42.32 |
3 |
3 |
1 |
45 |
472.693 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.43 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
97 |
0.34 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.43 |
4.57 |
-41.5 |
1 |
4 |
1 |
33 |
409.403 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
295 |
0.26 |
Binding ≤ 10μM
|
|
DPP4-1-E |
Dipeptidyl Peptidase IV (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
-4.84 |
-63.65 |
3 |
10 |
1 |
118 |
473.561 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.80 |
-3.99 |
-111.29 |
4 |
10 |
2 |
121 |
474.569 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
44 |
0.31 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
55 |
0.31 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
30 |
0.32 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
103 |
0.30 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
83 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.53 |
6.4 |
-90.09 |
0 |
3 |
2 |
7 |
451.743 |
16 |
↓
|
|
|
|
Analogs
-
5764620
-
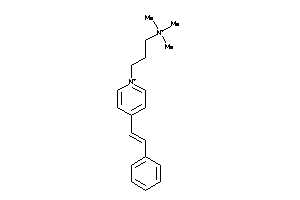
-
31320062
-
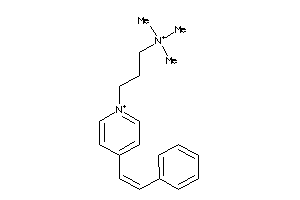
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
34 |
0.36 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
38 |
0.36 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
24 |
0.37 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
121 |
0.33 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
228 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.66 |
6.02 |
-90.39 |
0 |
3 |
2 |
7 |
395.635 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
8020 |
0.21 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
6280 |
0.21 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2830 |
0.23 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2470 |
0.23 |
Binding ≤ 10μM
|
|
SIRT1-1-E |
NAD-dependent Deacetylase Sirtuin 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
160 |
0.28 |
Binding ≤ 10μM
|
|
SIRT2-3-E |
NAD-dependent Deacetylase Sirtuin 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
160 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
8.12 |
-58.41 |
3 |
8 |
1 |
92 |
470.582 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.59 |
3.63 |
-91.33 |
9 |
9 |
2 |
152 |
424.484 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
12 |
0.32 |
Binding ≤ 10μM |
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
10.46 |
-51.7 |
3 |
8 |
1 |
87 |
479.649 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.90 |
12.58 |
-42.32 |
3 |
8 |
1 |
87 |
479.649 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.90 |
13.09 |
-186.7 |
5 |
8 |
3 |
90 |
481.665 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
12 |
0.32 |
Binding ≤ 10μM |
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
12.19 |
-48.28 |
3 |
8 |
1 |
87 |
479.649 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.90 |
10.88 |
-46.2 |
3 |
8 |
1 |
87 |
479.649 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.90 |
13.14 |
-180.81 |
5 |
8 |
3 |
90 |
481.665 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
35 |
0.45 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
81 |
0.43 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.38 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
260 |
0.40 |
Binding ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
1300 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
4.15 |
-11.62 |
2 |
5 |
0 |
55 |
308.385 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.50 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
22 |
0.47 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
22 |
0.47 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.42 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
55 |
0.44 |
Binding ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
64 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.56 |
-6.67 |
1 |
4 |
0 |
35 |
326.831 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
112 |
0.42 |
Binding ≤ 10μM |
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
112 |
0.42 |
Binding ≤ 10μM |
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
112 |
0.42 |
Binding ≤ 10μM |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
662 |
0.38 |
Binding ≤ 10μM |
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
662 |
0.38 |
Binding ≤ 10μM |
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
662 |
0.38 |
Binding ≤ 10μM |
|
ACM4-3-E |
Muscarinic Acetylcholine Receptor M4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
662 |
0.38 |
Binding ≤ 10μM |
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
662 |
0.38 |
Binding ≤ 10μM |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
266 |
0.40 |
Binding ≤ 10μM |
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
266 |
0.40 |
Binding ≤ 10μM |
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
266 |
0.40 |
Binding ≤ 10μM |
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
395 |
0.39 |
Binding ≤ 10μM |
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
87 |
0.43 |
Binding ≤ 10μM |
|
DRD2-4-E |
Dopamine D2 Receptor (cluster #4 Of 24), Eukaryotic |
Eukaryotes |
119 |
0.42 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
9.03 |
-17.08 |
1 |
5 |
0 |
49 |
328.441 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
14.32 |
-36.04 |
1 |
3 |
1 |
29 |
428.596 |
8 |
↓
|
|
|
|
Analogs
-
38606891
-
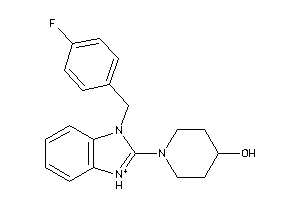
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
988 |
0.34 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6664 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
12.55 |
-91.76 |
3 |
4 |
2 |
39 |
340.446 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
19 |
0.39 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.32 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
10.55 |
-42.11 |
3 |
4 |
1 |
54 |
385.572 |
7 |
↓
|
|
|
|
Analogs
-
26153628
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
6559 |
0.25 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3441 |
0.26 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
950 |
0.29 |
Binding ≤ 10μM
|
|
ACM4-3-E |
Muscarinic Acetylcholine Receptor M4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
91 |
0.34 |
Binding ≤ 10μM
|
|
ACM5-3-E |
Muscarinic Acetylcholine Receptor M5 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7412 |
0.25 |
Binding ≤ 10μM
|
|
ACM4-2-E |
Muscarinic Acetylcholine Receptor M4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
7.45 |
-14.06 |
1 |
6 |
0 |
64 |
392.455 |
4 |
↓
|
|