|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.30 |
Binding ≤ 10μM
|
|
EAA2-1-E |
Excitatory Amino Acid Transporter 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.40 |
Binding ≤ 10μM
|
|
EAA3-1-E |
Excitatory Amino Acid Transporter 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
6.03 |
-34.53 |
4 |
6 |
0 |
106 |
415.19 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.01 |
5.72 |
-46.01 |
3 |
6 |
-1 |
104 |
414.182 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
40914195
-
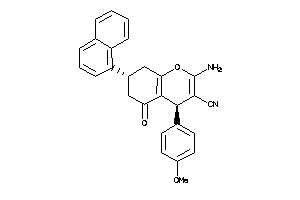
-
40914197
-
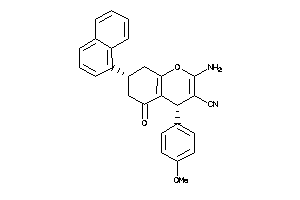
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
661 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.73 |
10.49 |
-15.71 |
2 |
5 |
0 |
85 |
422.484 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.73 |
10.87 |
-60.8 |
3 |
5 |
1 |
87 |
423.492 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6310 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
9.05 |
-15.36 |
2 |
4 |
0 |
76 |
342.398 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.99 |
9.5 |
-62.53 |
3 |
4 |
1 |
78 |
343.406 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6310 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
9.05 |
-15.3 |
2 |
4 |
0 |
76 |
342.398 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.99 |
9.51 |
-62.25 |
3 |
4 |
1 |
78 |
343.406 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6310 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
9.04 |
-15.32 |
2 |
4 |
0 |
76 |
342.398 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.99 |
9.5 |
-62.26 |
3 |
4 |
1 |
78 |
343.406 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6310 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
9.05 |
-15.36 |
2 |
4 |
0 |
76 |
342.398 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.99 |
9.51 |
-62.55 |
3 |
4 |
1 |
78 |
343.406 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
1.12 |
-15.29 |
2 |
5 |
0 |
85 |
372.424 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
8.39 |
-14.84 |
2 |
5 |
0 |
85 |
372.424 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
1.12 |
-14.34 |
2 |
5 |
0 |
85 |
372.424 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
1.12 |
-15.02 |
2 |
5 |
0 |
85 |
372.424 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3631 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
1.62 |
-16.09 |
2 |
7 |
0 |
121 |
387.395 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3631 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
1.62 |
-16.01 |
2 |
7 |
0 |
121 |
387.395 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3631 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
1.62 |
-15.76 |
2 |
7 |
0 |
121 |
387.395 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3631 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
1.62 |
-15.85 |
2 |
7 |
0 |
121 |
387.395 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2754 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
0.74 |
-15.04 |
2 |
4 |
0 |
76 |
388.492 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2754 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
0.74 |
-14.59 |
2 |
4 |
0 |
76 |
388.492 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2754 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
0.74 |
-13.86 |
2 |
4 |
0 |
76 |
388.492 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2754 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
0.74 |
-14.73 |
2 |
4 |
0 |
76 |
388.492 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7413 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.68 |
11.2 |
-15.6 |
2 |
4 |
0 |
76 |
392.458 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7413 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.68 |
1.25 |
-15.57 |
2 |
4 |
0 |
76 |
392.458 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7413 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.68 |
1.25 |
-15.55 |
2 |
4 |
0 |
76 |
392.458 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7413 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.68 |
1.24 |
-15.92 |
2 |
4 |
0 |
76 |
392.458 |
2 |
↓
|
|
|
|
Analogs
-
728135
-
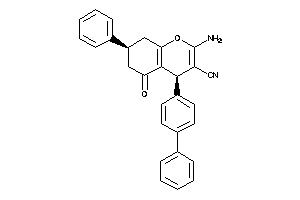
-
728136
-
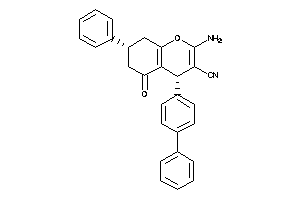
-
728137
-
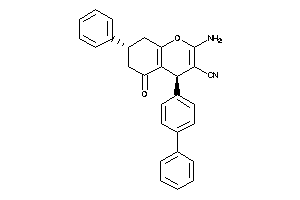
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2138 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
1.64 |
-15.01 |
2 |
4 |
0 |
76 |
418.496 |
3 |
↓
|
|
|
|
Analogs
-
728136
-

-
728137
-

-
728134
-
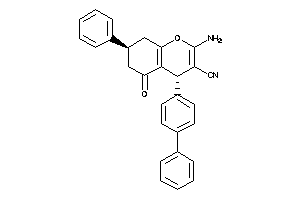
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2138 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
1.63 |
-15.16 |
2 |
4 |
0 |
76 |
418.496 |
3 |
↓
|
|
|
|
Analogs
-
728137
-

-
728134
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2138 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
1.64 |
-14.89 |
2 |
4 |
0 |
76 |
418.496 |
3 |
↓
|
|
|
|
Analogs
-
728134
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2138 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
1.64 |
-15.25 |
2 |
4 |
0 |
76 |
418.496 |
3 |
↓
|
|
|
|
Analogs
-
179162
-
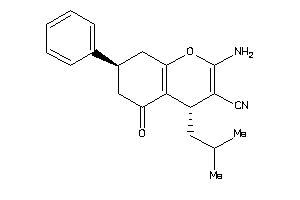
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9200 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
6.2 |
-11.73 |
2 |
4 |
0 |
76 |
280.327 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.67 |
6.58 |
-53.84 |
3 |
4 |
1 |
78 |
281.335 |
1 |
↓
|
|
|
|
Analogs
-
179162
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9200 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
6.2 |
-11.75 |
2 |
4 |
0 |
76 |
280.327 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.67 |
6.58 |
-53.65 |
3 |
4 |
1 |
78 |
281.335 |
1 |
↓
|
|
|
|
Analogs
-
179162
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9200 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
6.2 |
-11.77 |
2 |
4 |
0 |
76 |
280.327 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.67 |
6.58 |
-53.63 |
3 |
4 |
1 |
78 |
281.335 |
1 |
↓
|
|
|
|
Analogs
-
179162
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9200 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
6.2 |
-11.72 |
2 |
4 |
0 |
76 |
280.327 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.67 |
6.58 |
-53.79 |
3 |
4 |
1 |
78 |
281.335 |
1 |
↓
|
|
|
|
Analogs
-
659267
-
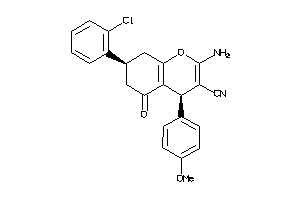
-
659270
-
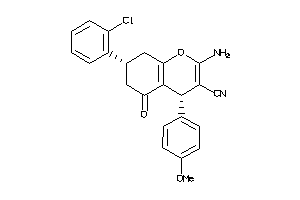
-
659271
-
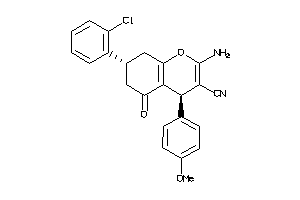
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3236 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.20 |
1 |
-15.47 |
2 |
5 |
0 |
85 |
406.869 |
3 |
↓
|
|
|
|
Analogs
-
659270
-

-
659271
-

-
659265
-
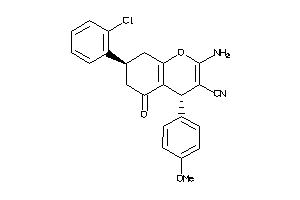
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3236 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.20 |
0.99 |
-16.04 |
2 |
5 |
0 |
85 |
406.869 |
3 |
↓
|
|
|
|
Analogs
-
659271
-

-
659265
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3236 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.20 |
1 |
-16.02 |
2 |
5 |
0 |
85 |
406.869 |
3 |
↓
|
|
|
|
Analogs
-
659265
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3236 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.20 |
0.99 |
-16.01 |
2 |
5 |
0 |
85 |
406.869 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7413 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
1.57 |
-13.67 |
2 |
5 |
0 |
89 |
332.359 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7413 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
1.57 |
-13.58 |
2 |
5 |
0 |
89 |
332.359 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7413 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
1.57 |
-13.8 |
2 |
5 |
0 |
89 |
332.359 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7413 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
1.57 |
-13.84 |
2 |
5 |
0 |
89 |
332.359 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.48 |
Binding ≤ 10μM
|
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3162 |
0.45 |
Binding ≤ 10μM
|
|
EAA2-1-E |
Excitatory Amino Acid Transporter 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.45 |
Binding ≤ 10μM
|
|
EAA2-1-E |
Excitatory Amino Acid Transporter 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
EAA3-1-E |
Excitatory Amino Acid Transporter 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
EAA3-1-E |
Excitatory Amino Acid Transporter 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
EAA3-1-E |
Excitatory Amino Acid Transporter 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9300 |
0.41 |
Binding ≤ 10μM
|
|
EAA4-1-E |
Excitatory Amino Acid Transporter 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.48 |
Binding ≤ 10μM
|
|
EAA5-1-E |
Excitatory Amino Acid Transporter 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.48 |
Binding ≤ 10μM
|
|
SATT-1-E |
Neutral Amino Acid Transporter A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.22 |
4.82 |
-55.21 |
3 |
6 |
-1 |
117 |
238.219 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.22 |
2.86 |
-35.81 |
4 |
6 |
0 |
114 |
239.227 |
6 |
↓
|
|
|
|
Analogs
-
1176209
-
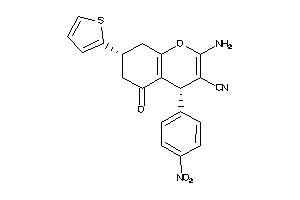
-
1176210
-
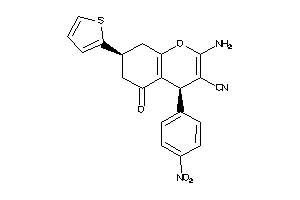
-
1176211
-
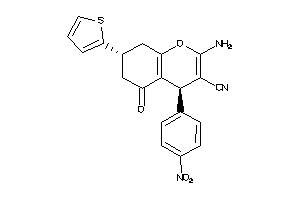
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9900 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
0.75 |
-15.24 |
2 |
7 |
0 |
121 |
393.424 |
3 |
↓
|
|
|
|
Analogs
-
1176210
-

-
1176211
-

-
1176208
-
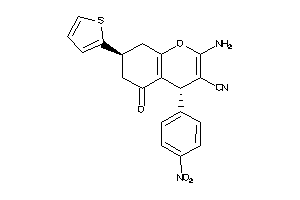
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9900 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
0.76 |
-15.35 |
2 |
7 |
0 |
121 |
393.424 |
3 |
↓
|
|
|
|
Analogs
-
1176211
-

-
1176208
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9900 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
0.76 |
-15.26 |
2 |
7 |
0 |
121 |
393.424 |
3 |
↓
|
|
|
|
Analogs
-
1176208
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9900 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
0.76 |
-15.42 |
2 |
7 |
0 |
121 |
393.424 |
3 |
↓
|
|
|
|
Analogs
-
706102
-
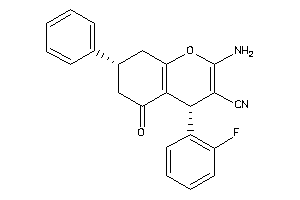
-
706103
-
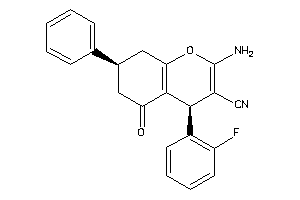
-
706104
-
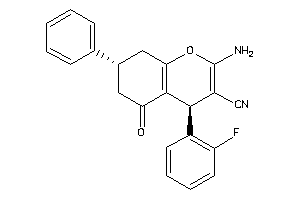
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4786 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
1.8 |
-17.89 |
2 |
4 |
0 |
76 |
360.388 |
2 |
↓
|
|
|
|
Analogs
-
706103
-

-
706104
-

-
706101
-
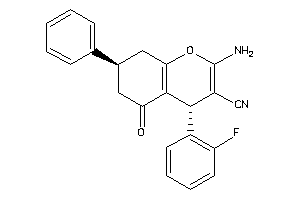
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4786 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
1.8 |
-17.55 |
2 |
4 |
0 |
76 |
360.388 |
2 |
↓
|
|
|
|
Analogs
-
706104
-

-
706101
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4786 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
1.81 |
-17.55 |
2 |
4 |
0 |
76 |
360.388 |
2 |
↓
|
|
|
|
Analogs
-
706101
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EAA1-1-E |
Excitatory Amino Acid Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4786 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
1.8 |
-17.59 |
2 |
4 |
0 |
76 |
360.388 |
2 |
↓
|
|