|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.31 |
Binding ≤ 10μM
|
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.30 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
-0.07 |
-69.14 |
0 |
8 |
-1 |
102 |
527.597 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.42 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
AA1R-3-E |
Adenosine A1 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
980 |
0.38 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2800 |
0.35 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1400 |
0.37 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4500 |
0.34 |
Binding ≤ 10μM
|
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.49 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
65 |
0.46 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.47 |
Binding ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
530 |
0.40 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
840 |
0.39 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
AOFA-1-E |
Monoamine Oxidase A (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
GASR-1-E |
Cholecystokinin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
GCR-1-E |
Glucocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
610 |
0.40 |
Binding ≤ 10μM
|
|
HRH2-3-E |
Histamine H2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
HRH3-3-E |
Histamine H3 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
MC4R-1-E |
Melanocortin Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
MK14-2-E |
MAP Kinase P38 Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
MT3-1-E |
Metallothionein-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
NISCH-2-E |
Nischarin (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
NK1R-1-E |
Neurokinin 1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
NK2R-1-E |
Neurokinin 2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
NPY1R-2-E |
Neuropeptide Y Receptor Type 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
NTR1-1-E |
Neurotensin Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
OPRK-4-E |
Kappa Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
SC5A7-2-E |
High Affinity Choline Transporter 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.43 |
Functional ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Binding ≤ 10μM
|
|
Z104283-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #2 Of 4), Other |
Other |
1000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.33 |
2.67 |
-39.39 |
1 |
2 |
1 |
21 |
302.482 |
8 |
↓
|
|
|
|
Analogs
-
4321884
-
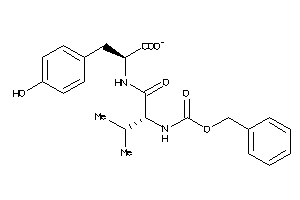
-
4321885
-
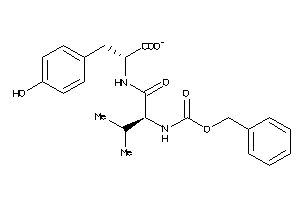
-
4321886
-
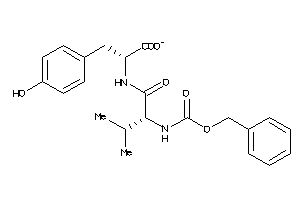
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
6.7 |
-50.08 |
3 |
8 |
-1 |
128 |
413.45 |
10 |
↓
|
|
|
|
|
|
|
Analogs
-
4321886
-

-
4321884
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
6.73 |
-53.67 |
3 |
8 |
-1 |
128 |
413.45 |
10 |
↓
|
|
|
|
Analogs
-
4321884
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
6.77 |
-51.32 |
3 |
8 |
-1 |
128 |
413.45 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.38 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
-2.52 |
-56.15 |
0 |
8 |
-1 |
102 |
409.418 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
2.85 |
-68 |
0 |
9 |
-1 |
112 |
505.499 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
10.47 |
-23 |
1 |
9 |
0 |
120 |
461.426 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
10.42 |
-16.84 |
1 |
9 |
0 |
120 |
461.426 |
6 |
↓
|
|
|
|
Analogs
-
3644203
-
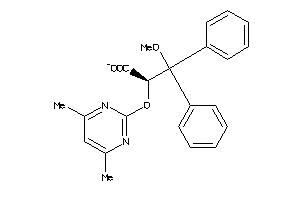
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
8.98 |
-62.91 |
0 |
6 |
-1 |
84 |
377.42 |
7 |
↓
|
|
|
|
Analogs
-
538627
-
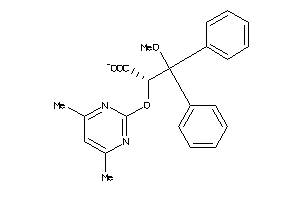
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
195 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
8.97 |
-64.38 |
0 |
6 |
-1 |
84 |
377.42 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
19 |
0.40 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
8.44 |
-62.47 |
0 |
6 |
-1 |
84 |
363.393 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
19 |
0.40 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
8.38 |
-62.22 |
0 |
6 |
-1 |
84 |
363.393 |
7 |
↓
|
|
|
|
Analogs
-
13140305
-
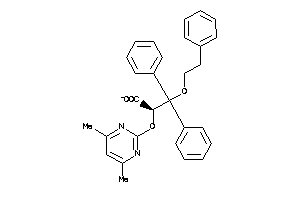
-
4675135
-
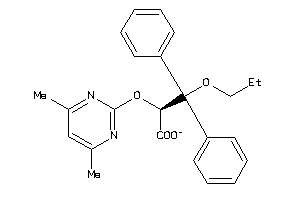
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.30 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
13.9 |
-67.77 |
0 |
6 |
-1 |
84 |
467.545 |
10 |
↓
|
|
|
|
Analogs
-
9660252
-
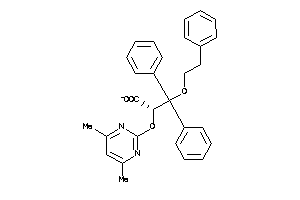
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.30 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
13.91 |
-67.98 |
0 |
6 |
-1 |
84 |
467.545 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.26 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.23 |
14.43 |
-59.58 |
3 |
11 |
-1 |
148 |
603.744 |
12 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.23 |
13.51 |
-59.16 |
4 |
11 |
0 |
150 |
604.752 |
12 |
↓
|
|
|
|
Analogs
-
34026477
-
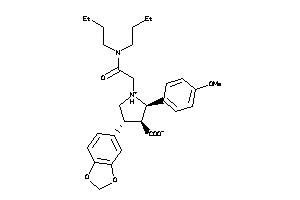
-
3812144
-
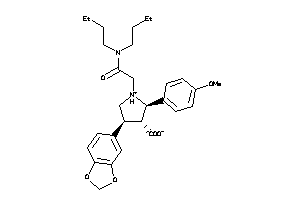
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
13.4 |
-57.6 |
1 |
8 |
0 |
93 |
510.631 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.64 |
11.89 |
-56.97 |
0 |
8 |
-1 |
91 |
509.623 |
12 |
↓
|
|
|
|
Analogs
-
34026476
-
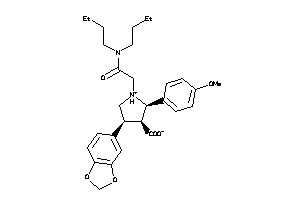
-
3812144
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
13.81 |
-58.85 |
1 |
8 |
0 |
93 |
510.631 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.64 |
12.1 |
-59.51 |
0 |
8 |
-1 |
91 |
509.623 |
12 |
↓
|
|
|
|
Analogs
-
13740975
-
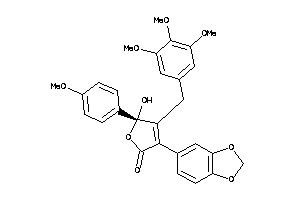
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.23 |
Binding ≤ 10μM
|
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
EDNRB-1-E |
Endothelin Receptor ET-B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
480 |
0.24 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
1 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
8.93 |
-20.37 |
1 |
9 |
0 |
102 |
506.507 |
8 |
↓
|
|
|
|
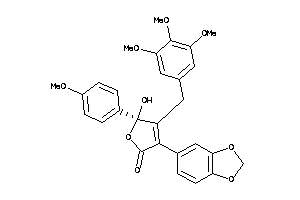
Analogs
-
13740974
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.23 |
Binding ≤ 10μM
|
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
EDNRB-1-E |
Endothelin Receptor ET-B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
480 |
0.24 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
1 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
8.88 |
-20.54 |
1 |
9 |
0 |
102 |
506.507 |
8 |
↓
|
|
|
|
Analogs
-
34026476
-

-
34026477
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
1.68 |
-51.98 |
1 |
8 |
0 |
92 |
510.631 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Lo
Low (pH 4.5-6)
|
5.45 |
12.76 |
-51.26 |
2 |
8 |
0 |
119 |
531.605 |
10 |
↓
|
|
|
|
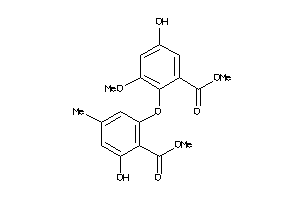
Analogs
-
605688
-
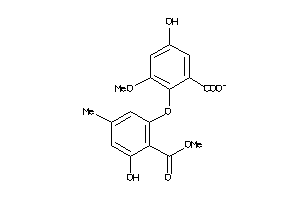
-
39560000
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
5.06 |
-75.7 |
2 |
8 |
-1 |
125 |
347.299 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.05 |
5.82 |
-116.91 |
1 |
8 |
-2 |
128 |
346.291 |
6 |
↓
|
|