|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3100 |
0.23 |
Binding ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
329 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
-1.89 |
-17.12 |
2 |
9 |
0 |
111 |
459.506 |
5 |
↓
|
|
|
|
Analogs
-
4426033
-
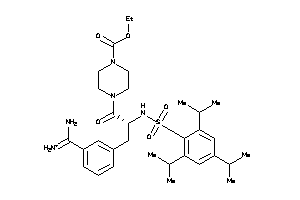
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
10.74 |
-41.24 |
5 |
10 |
1 |
148 |
614.833 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.37 |
Binding ≤ 10μM
|
|
FA9-1-E |
Coagulation Factor IX (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.23 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1200 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.11 |
5.22 |
-21.63 |
2 |
8 |
0 |
107 |
479.986 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
1.18 |
4.09 |
-51.74 |
3 |
8 |
1 |
112 |
480.994 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.35 |
Binding ≤ 10μM
|
|
O77669-1-E |
Coagulation Factor X (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
-4.37 |
-84.32 |
5 |
8 |
1 |
121 |
410.498 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
26 |
0.30 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
7.05 |
-77.8 |
6 |
10 |
0 |
154 |
477.569 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.50 |
7.51 |
-87.34 |
7 |
10 |
1 |
155 |
478.577 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.50 |
8.32 |
-110.06 |
8 |
10 |
2 |
160 |
479.585 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.12 |
4.51 |
-51.08 |
7 |
12 |
1 |
189 |
564.714 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
-5.37 |
-127.46 |
7 |
9 |
1 |
140 |
501.611 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
9.88 |
-80.97 |
6 |
8 |
2 |
127 |
521.687 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.16 |
9.11 |
-24.65 |
1 |
7 |
0 |
101 |
573.067 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
7.09 |
-12 |
2 |
7 |
0 |
72 |
459.594 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.57 |
9.47 |
-46.44 |
3 |
7 |
1 |
73 |
460.602 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.57 |
11.62 |
-117.36 |
4 |
7 |
2 |
74 |
461.61 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
6.54 |
-22.63 |
1 |
8 |
0 |
114 |
538.072 |
7 |
↓
|
|
|
|
Analogs
-
14113515
-
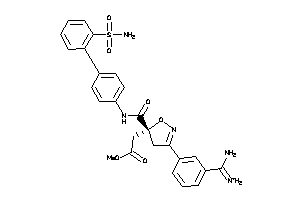
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3100 |
0.20 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
98 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
5.3 |
-48.98 |
7 |
11 |
1 |
189 |
536.59 |
9 |
↓
|
|
|
|
Analogs
-
3920966
-
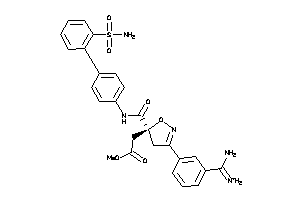
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3100 |
0.20 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
98 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
5.28 |
-51.08 |
7 |
11 |
1 |
189 |
536.59 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
3.62 |
-47.9 |
7 |
9 |
1 |
167 |
462.511 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.31 |
Binding ≤ 10μM
|
|
TRY2-1-E |
Trypsin II (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
9.49 |
-89.29 |
6 |
8 |
2 |
127 |
521.687 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
4.68 |
-44.4 |
7 |
9 |
1 |
159 |
475.554 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
TRY2-1-E |
Trypsin II (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
28 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
-3.75 |
-84.96 |
6 |
8 |
2 |
119 |
473.577 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.34 |
Binding ≤ 10μM
|
|
TRY2-1-E |
Trypsin II (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
21 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
7.58 |
-93.79 |
6 |
8 |
2 |
127 |
501.678 |
8 |
↓
|
|
|
|
Analogs
-
36751288
-
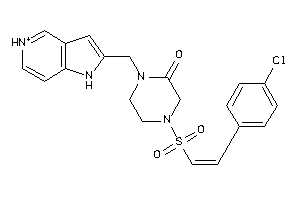
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
48 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
-5.5 |
-16.3 |
1 |
7 |
0 |
86 |
430.917 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.38 |
-5.39 |
-44.55 |
2 |
7 |
1 |
88 |
431.925 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
5.39 |
-21.79 |
3 |
9 |
0 |
117 |
482.562 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
-1.53 |
-64.17 |
4 |
7 |
1 |
108 |
533.527 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
|
THRB-1-E |
Thrombin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
11.22 |
-49.82 |
3 |
7 |
1 |
80 |
454.982 |
6 |
↓
|
|
|
|
Analogs
-
34025326
-
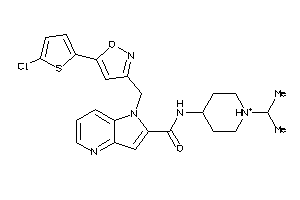
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
THRB-4-E |
Prothrombin (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
2760 |
0.24 |
Binding ≤ 10μM
|
|
THRB-1-E |
Thrombin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.58 |
13.43 |
-41.46 |
2 |
6 |
1 |
65 |
484.045 |
6 |
↓
|
|
|
|
Analogs
-
1494180
-
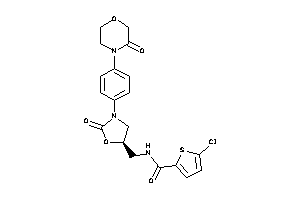
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
-3.62 |
-20.72 |
1 |
8 |
0 |
88 |
435.889 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
-6.86 |
-92.71 |
6 |
10 |
1 |
167 |
538.65 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.29 |
Binding ≤ 10μM
|
|
Z102352-1-O |
Plasma (cluster #1 Of 3), Other |
Other |
4400 |
0.19 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
10.24 |
-47.01 |
5 |
9 |
1 |
130 |
540.554 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.33 |
7.74 |
-19.4 |
4 |
9 |
0 |
129 |
539.546 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.32 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.59 |
-9.99 |
-74.27 |
7 |
12 |
1 |
190 |
546.655 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.34 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.05 |
3.88 |
-50.81 |
6 |
12 |
1 |
180 |
557.078 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.33 |
Binding ≤ 10μM
|
|
KLK1-1-E |
Kallikrein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9700 |
0.19 |
Binding ≤ 10μM
|
|
THRB-4-E |
Prothrombin (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.70 |
16.5 |
-37.08 |
2 |
5 |
1 |
51 |
514.024 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
3.44 |
-38.88 |
3 |
6 |
1 |
75 |
488.44 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
4.78 |
-43.04 |
5 |
8 |
1 |
127 |
453.544 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
4.95 |
-18.01 |
2 |
8 |
0 |
97 |
451.932 |
7 |
↓
|
|
|
|
Analogs
-
3964126
-
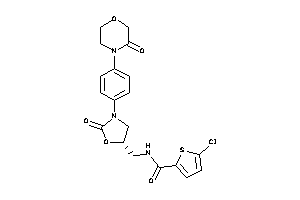
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
5.78 |
-22.12 |
1 |
8 |
0 |
88 |
435.889 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3601 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
12.72 |
-54.29 |
3 |
5 |
1 |
63 |
473.062 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.19 |
2.87 |
-78.93 |
6 |
11 |
0 |
202 |
547.611 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.19 |
4.48 |
-99.1 |
5 |
11 |
-1 |
204 |
546.603 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
-3.76 |
-107.21 |
7 |
10 |
1 |
162 |
557.675 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
301 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.17 |
-0.54 |
-64.82 |
5 |
8 |
1 |
132 |
447.515 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.97 |
-4.39 |
-75.96 |
5 |
10 |
0 |
154 |
471.521 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
11 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
-0.06 |
-13.98 |
2 |
4 |
0 |
58 |
438.911 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.38 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8900 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
8.66 |
-41.9 |
4 |
8 |
1 |
109 |
452.922 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.26 |
Binding ≤ 10μM
|
|
FA12-1-E |
Coagulation Factor XII (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
180 |
0.30 |
Binding ≤ 10μM
|
|
KLKB1-3-E |
Plasma Kallikrein (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
31 |
0.34 |
Binding ≤ 10μM
|
|
TF-1-E |
Coagulation Factor III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.26 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2500 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.34 |
5.3 |
-88.32 |
9 |
9 |
2 |
156 |
452.987 |
13 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.34 |
3.86 |
-48.36 |
8 |
9 |
1 |
151 |
451.979 |
13 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.29 |
3.81 |
-67.75 |
8 |
9 |
1 |
164 |
451.979 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.58 |
-0.95 |
-13.4 |
2 |
4 |
0 |
58 |
455.366 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
960 |
0.27 |
Binding ≤ 10μM
|
|
FA9-1-E |
Coagulation Factor IX (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
29 |
0.34 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
250 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
8.39 |
-38.54 |
6 |
6 |
1 |
102 |
431.541 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
960 |
0.27 |
Binding ≤ 10μM
|
|
FA9-1-E |
Coagulation Factor IX (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
29 |
0.34 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
250 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
8.46 |
-39.61 |
6 |
6 |
1 |
102 |
431.541 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
THRB-4-E |
Prothrombin (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
2000 |
0.21 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
280 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.41 |
-5.19 |
-105.82 |
6 |
11 |
1 |
163 |
527.508 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
96 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
-2.56 |
-40.01 |
5 |
5 |
1 |
85 |
447.562 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.33 |
Binding ≤ 10μM
|
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.25 |
Binding ≤ 10μM
|
|
PROC-1-E |
Vitamin K-dependent Protein C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.24 |
Binding ≤ 10μM
|
|
TRY2-1-E |
Trypsin II (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
5.49 |
-40.89 |
5 |
7 |
1 |
118 |
423.518 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANT3-1-E |
Antithrombin-III Precursor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.24 |
Binding ≤ 10μM
|
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7900 |
0.19 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
7 |
0.31 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
690 |
0.23 |
Binding ≤ 10μM |
|
TRY2-1-E |
Trypsin II (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
690 |
0.23 |
Binding ≤ 10μM
|
|
TRY3-1-E |
Trypsin III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
690 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.23 |
6.47 |
-52.48 |
6 |
9 |
1 |
147 |
522.651 |
9 |
↓
|
|