|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.26 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.36 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.35 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8220 |
0.21 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.26 |
Binding ≤ 10μM
|
|
SSR5-1-E |
Somatostatin Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4070 |
0.22 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.27 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.24 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3300 |
0.23 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
734 |
0.25 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
8000 |
0.21 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
6500 |
0.21 |
Functional ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
10 |
0.33 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
1 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.71 |
17.29 |
-53.86 |
2 |
5 |
1 |
44 |
459.589 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.71 |
17.71 |
-105.8 |
3 |
5 |
2 |
45 |
460.597 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.41 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.41 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
99 |
0.34 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
92 |
0.34 |
Functional ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
92 |
0.34 |
Functional ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
92 |
0.34 |
Functional ≤ 10μM
|
|
Z80401-1-O |
PSN1 (cluster #1 Of 2), Other |
Other |
92 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.95 |
11.33 |
-19.72 |
0 |
6 |
0 |
65 |
405.889 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.95 |
11.85 |
-53.13 |
1 |
6 |
1 |
66 |
406.897 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.33 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.29 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
|
Q5EI73-1-E |
Protein Farnesyltransferase Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM
|
|
Q5EI74-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.29 |
Functional ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.29 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4060 |
0.22 |
ADME/T ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
4 |
0.35 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
2 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
13.62 |
-14.28 |
2 |
5 |
0 |
66 |
489.406 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.09 |
14.08 |
-48.31 |
3 |
5 |
1 |
67 |
490.414 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.09 |
14.29 |
-131.87 |
4 |
5 |
2 |
69 |
491.422 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.34 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.34 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.27 |
Functional ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.27 |
Functional ≤ 10μM
|
|
RASH-1-E |
Transforming Protein P21/H-Ras-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.28 |
Functional ≤ 10μM
|
|
Z80106-1-O |
COS-1 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
75 |
0.28 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
50 |
0.28 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
70 |
0.28 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
160 |
0.26 |
Functional ≤ 10μM
|
|
Z81285-1-O |
NIH-H Cell Line (cluster #1 Of 1), Other |
Other |
72 |
0.28 |
Functional ≤ 10μM
|
|
Z81286-1-O |
NIH-K Cell Line (cluster #1 Of 1), Other |
Other |
500 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.84 |
-3.64 |
-16.37 |
2 |
6 |
0 |
79 |
638.832 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.84 |
-3.78 |
-49.07 |
3 |
6 |
1 |
80 |
639.84 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.29 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.29 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.29 |
Functional ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.29 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
16 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
1.87 |
-87.5 |
2 |
5 |
0 |
73 |
538.798 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.43 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.43 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.29 |
Functional ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.29 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
1000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.47 |
8.41 |
-93.2 |
5 |
6 |
0 |
109 |
433.599 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.47 |
9.03 |
-87.61 |
5 |
6 |
-1 |
109 |
432.591 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.47 |
8.08 |
-57.51 |
4 |
6 |
-1 |
107 |
432.591 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.29 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
21 |
0.35 |
Binding ≤ 10μM |
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
21 |
0.35 |
Binding ≤ 10μM |
|
Q6IE76-1-E |
Squalene Synthetase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
21 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.35 |
7.28 |
-68.22 |
6 |
7 |
0 |
124 |
470.705 |
16 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.35 |
8.42 |
-109.32 |
7 |
7 |
0 |
126 |
470.705 |
16 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.35 |
7.44 |
-75.79 |
6 |
7 |
0 |
121 |
470.705 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.35 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
8.54 |
-58.47 |
5 |
6 |
1 |
95 |
448.634 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.54 |
8.11 |
-13.19 |
4 |
6 |
0 |
93 |
447.626 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.54 |
9.08 |
-45.85 |
5 |
6 |
0 |
95 |
447.626 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5400 |
0.22 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5400 |
0.22 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
38 |
0.31 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Functional ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
13.02 |
-90.01 |
4 |
7 |
0 |
111 |
456.546 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.93 |
12.59 |
-64.77 |
3 |
7 |
-1 |
110 |
455.538 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.93 |
12.47 |
-64.66 |
3 |
7 |
-1 |
110 |
455.538 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
55 |
0.31 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.31 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
55 |
0.31 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Functional ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
9.62 |
-98.2 |
5 |
6 |
0 |
109 |
465.619 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.30 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.28 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
54 |
0.30 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.28 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.22 |
Functional ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
10.59 |
-58.96 |
5 |
6 |
1 |
95 |
480.654 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.56 |
10.67 |
-41.15 |
5 |
6 |
0 |
95 |
479.646 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.56 |
9.82 |
-10.1 |
4 |
6 |
0 |
93 |
479.646 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.40 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.40 |
Binding ≤ 10μM
|
|
ILVB-1-F |
Acetolactate Synthase Catalytic Subunit, Mitochondrial (cluster #1 Of 1), Fungal |
Fungi |
200 |
0.45 |
Binding ≤ 10μM
|
|
Z50420-2-O |
Trypanosoma Brucei Brucei (cluster #2 Of 7), Other |
Other |
10 |
0.53 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
3600 |
0.36 |
Functional ≤ 10μM
|
|
Z50442-2-O |
Candida Albicans (cluster #2 Of 4), Other |
Other |
3100 |
0.37 |
Functional ≤ 10μM
|
|
Z50476-1-O |
Tetrahymena Pyriformis (cluster #1 Of 1), Other |
Other |
380 |
0.43 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
3 |
0.57 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
150 |
0.45 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
2450 |
0.37 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 5), Other |
Other |
123 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 2), Other |
Other |
1000 |
0.40 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.47 |
1.87 |
-13.46 |
2 |
6 |
0 |
81 |
326.399 |
1 |
↓
|
|
|
|
Analogs
-
602601
-
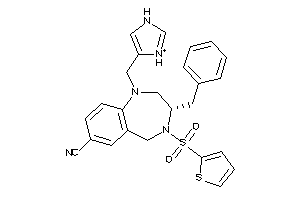
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.23 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.23 |
Binding ≤ 10μM
|
|
Q5EI73-1-E |
Protein Farnesyltransferase Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.33 |
Binding ≤ 10μM
|
|
Q5EI74-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.33 |
Binding ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
25 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
12.08 |
-52.1 |
2 |
7 |
1 |
94 |
490.634 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
3.66 |
12.06 |
-52.02 |
2 |
7 |
1 |
94 |
490.634 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.66 |
11.8 |
-15.02 |
1 |
7 |
0 |
93 |
489.626 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.28 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
9.59 |
-59.01 |
5 |
6 |
1 |
95 |
480.654 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.56 |
10.06 |
-43.94 |
5 |
6 |
0 |
95 |
479.646 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.56 |
9.26 |
-10.11 |
4 |
6 |
0 |
93 |
479.646 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.28 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
10.65 |
-66.46 |
5 |
6 |
1 |
95 |
480.654 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.56 |
11.12 |
-51.68 |
5 |
6 |
0 |
95 |
479.646 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.56 |
10.19 |
-16.5 |
4 |
6 |
0 |
93 |
479.646 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.28 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
10.66 |
-66.76 |
5 |
6 |
1 |
95 |
480.654 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.56 |
11.14 |
-51.98 |
5 |
6 |
0 |
95 |
479.646 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.56 |
10.21 |
-15.9 |
4 |
6 |
0 |
93 |
479.646 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.40 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.12 |
-5.54 |
-90.98 |
5 |
6 |
0 |
108 |
407.561 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.12 |
-6.5 |
-89.59 |
5 |
6 |
-1 |
108 |
406.553 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.42 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.93 |
3.06 |
-90.54 |
5 |
7 |
0 |
126 |
371.484 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.93 |
3.59 |
-82.89 |
5 |
7 |
-1 |
126 |
370.476 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.30 |
Binding ≤ 10μM |
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.30 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.53 |
6.92 |
-84.57 |
6 |
9 |
0 |
155 |
548.731 |
14 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.53 |
6.59 |
-54.76 |
5 |
9 |
-1 |
153 |
547.723 |
14 |
↓
|
|