|
|
Analogs
-
120294
-
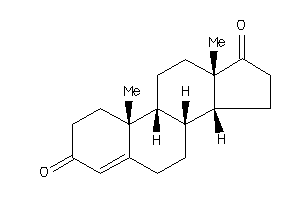
-
2046798
-
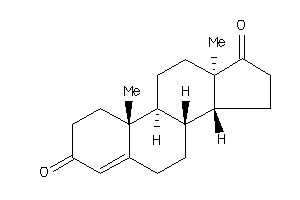
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Binding ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
FABPL-1-E |
Fatty Acid-binding Protein, Liver (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.46 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
31 |
0.46 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.48 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
SGMR1-2-E |
Sigma Opioid Receptor (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
260 |
0.40 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.30 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.30 |
Functional ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2770 |
0.34 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Functional ≤ 10μM
|
|
CP2C9-2-E |
Cytochrome P450 2C9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5500 |
0.32 |
ADME/T ≤ 10μM
|
|
ERG2-1-F |
C-8 Sterol Isomerase (cluster #1 Of 2), Fungal |
Fungi |
4430 |
0.33 |
Binding ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
6310 |
0.32 |
Functional ≤ 10μM
|
|
Z80712-1-O |
T47D (Breast Carcinoma Cells) (cluster #1 Of 7), Other |
Other |
1 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
10.64 |
-9.9 |
0 |
2 |
0 |
34 |
314.469 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.39 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.39 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1197 |
0.30 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.40 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.40 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Functional ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
924 |
0.30 |
Functional ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Functional ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1197 |
0.30 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1197 |
0.30 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
11 |
0.40 |
Binding ≤ 10μM
|
|
Z80712-1-O |
T47D (Breast Carcinoma Cells) (cluster #1 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80712-1-O |
T47D (Breast Carcinoma Cells) (cluster #1 Of 7), Other |
Other |
1 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
11.72 |
-14.72 |
0 |
4 |
0 |
60 |
386.532 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.62 |
2.72 |
-14.93 |
3 |
5 |
0 |
95 |
362.466 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
-
-
-
- API0002946
- API0002947
- API0005043
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
|
Z50594-7-O |
Mus Musculus (cluster #7 Of 9), Other |
Other |
15 |
0.42 |
Functional ≤ 10μM |
|
Z80418-3-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #3 Of 9), Other |
Other |
61 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.62 |
3.13 |
-14.6 |
3 |
5 |
0 |
95 |
362.466 |
2 |
↓
|
|
|
|
Analogs
-
3814382
-
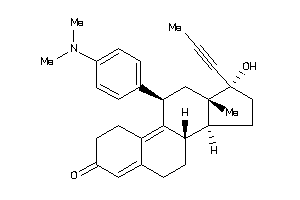
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
89 |
0.31 |
Binding ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.23 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2876 |
0.24 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
410 |
0.28 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
640 |
0.27 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.37 |
Binding ≤ 10μM |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.35 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2244 |
0.25 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
812 |
0.27 |
Functional ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.38 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1155 |
0.26 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.37 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4700 |
0.23 |
ADME/T ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
6310 |
0.23 |
Functional ≤ 10μM
|
|
Z80110-2-O |
CV-1 (Kidney Cells) (cluster #2 Of 2), Other |
Other |
10 |
0.35 |
Functional ≤ 10μM
|
|
Z80491-2-O |
SK-N-MC (Neuroepithelioma Cells) (cluster #2 Of 4), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
2 |
0.38 |
Functional ≤ 10μM
|
|
VP16-1-V |
Alpha Trans-inducing Protein (VP16) (cluster #1 Of 1), Viral |
Viruses |
1 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.60 |
1.26 |
-10.46 |
1 |
3 |
0 |
40 |
429.604 |
2 |
↓
|
|
|
|
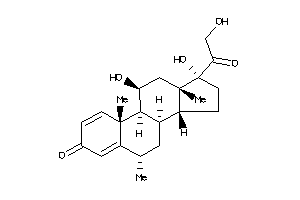
Analogs
-
3881944
-
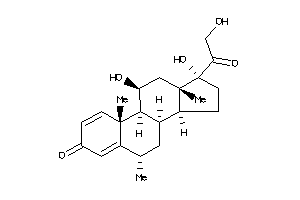
-
3881945
-
-
3881946
-
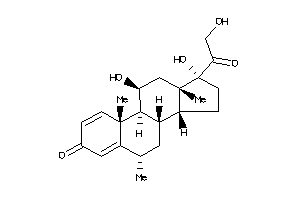
-
5275964
-
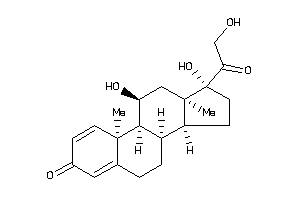
-
5276791
-
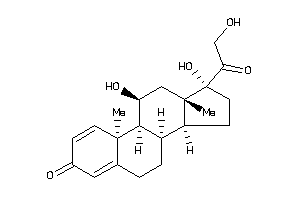
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2762 |
0.30 |
Binding ≤ 10μM
|
|
FABPL-1-E |
Fatty Acid-binding Protein, Liver (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2660 |
0.30 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
GLNA-1-E |
Glutamine Synthetase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.40 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
37 |
0.40 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.41 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM |
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
25 |
0.41 |
Functional ≤ 10μM
|
|
Z50594-7-O |
Mus Musculus (cluster #7 Of 9), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM
|
|
Z80110-2-O |
CV-1 (Kidney Cells) (cluster #2 Of 2), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
760 |
0.33 |
Functional ≤ 10μM
|
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
5 |
0.45 |
Functional ≤ 10μM
|
|
Z80954-3-O |
HFF (Foreskin Fibroblasts) (cluster #3 Of 4), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z81011-3-O |
Human Cell Lines (cluster #3 Of 3), Other |
Other |
48 |
0.39 |
Functional ≤ 10μM
|
|
Z81247-4-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #4 Of 9), Other |
Other |
16 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
3.09 |
-16.14 |
3 |
5 |
0 |
95 |
360.45 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4R)-N-cyclopropyl-4-[(3R,5S,7S,8R,9S,10S,13R,14S,17R)-3,7-dihydroxy-10,13-dimethyl-2,3,4,5,6,7,8,9,
(4R)-N-cyclopropyl-4-[(3R,5S,7S,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7300 |
0.23 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.47 |
6.59 |
-10.35 |
3 |
4 |
0 |
70 |
431.661 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4R)-1-(4-benzylpiperazin-1-yl)-4-[(3R,5S,7S,8R,9S,10S,13R,14S,17R)-3,7-dihydroxy-10,13-dimethyl-2,3
(4R)-1-(4-benzylpiperazin-1-yl)-…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5600 |
0.18 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
10.06 |
-11.91 |
2 |
5 |
0 |
64 |
550.828 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.64 |
12.27 |
-50.51 |
3 |
5 |
1 |
65 |
551.836 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
4026402
-
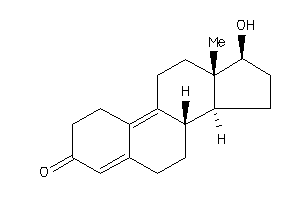
-
4026403
-
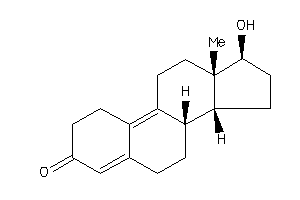
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
133 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
-1.09 |
-8.22 |
1 |
2 |
0 |
37 |
272.388 |
0 |
↓
|
|
|
|
Analogs
-
4026402
-

-
4026403
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
133 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
-1.01 |
-7.98 |
1 |
2 |
0 |
37 |
272.388 |
0 |
↓
|
|
|
|
Analogs
-
4475234
-
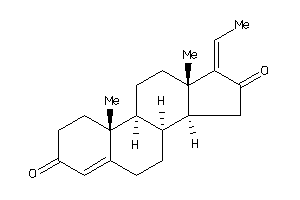
-
4475235
-
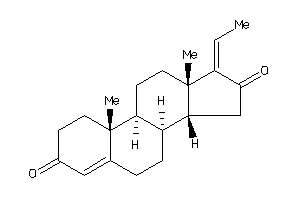
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
240 |
0.40 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
224 |
0.40 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.45 |
Binding ≤ 10μM
|
|
NR1I2-2-E |
Pregnane X Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
201 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
10.56 |
-10.66 |
0 |
2 |
0 |
34 |
312.453 |
0 |
↓
|
|
|
|
Analogs
-
4475235
-

-
4475232
-
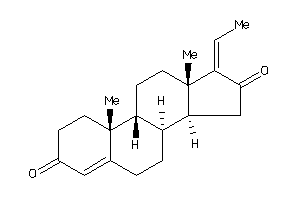
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
240 |
0.40 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
224 |
0.40 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.45 |
Binding ≤ 10μM
|
|
NR1I2-2-E |
Pregnane X Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
201 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
10.59 |
-10.36 |
0 |
2 |
0 |
34 |
312.453 |
0 |
↓
|
|
|
|
Analogs
-
4475232
-

-
4475234
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
240 |
0.40 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
224 |
0.40 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.45 |
Binding ≤ 10μM
|
|
NR1I2-2-E |
Pregnane X Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
201 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
10.37 |
-10.88 |
0 |
2 |
0 |
34 |
312.453 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
240 |
0.40 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
224 |
0.40 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.45 |
Binding ≤ 10μM
|
|
NR1I2-2-E |
Pregnane X Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
201 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
10.65 |
-12.38 |
0 |
2 |
0 |
34 |
312.453 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.35 |
Binding ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1900 |
0.24 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
85 |
0.30 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.25 |
Binding ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.54 |
0.4 |
-14.47 |
1 |
5 |
0 |
68 |
449.591 |
5 |
↓
|
|
|
|
Analogs
-
3874177
-
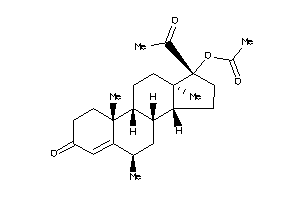
-
3874178
-
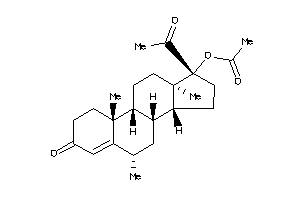
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
3.87 |
-14.13 |
0 |
4 |
0 |
60 |
386.532 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
36584219
-
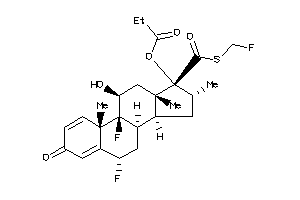
-
39741203
-
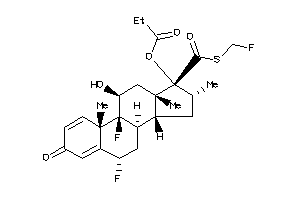
-
3920027
-
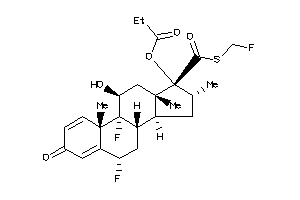
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.61 |
10.73 |
-22.95 |
1 |
5 |
0 |
81 |
500.579 |
6 |
↓
|
|
|
|
Analogs
-
3920027
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.61 |
11.05 |
-13.07 |
1 |
5 |
0 |
81 |
500.579 |
6 |
↓
|
|
|
|
Analogs
-
3920027
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.61 |
10.59 |
-15.56 |
1 |
5 |
0 |
81 |
500.579 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
3978012
-
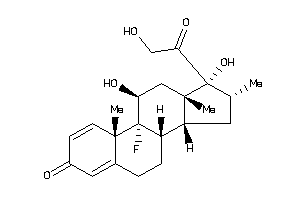
-
3978013
-
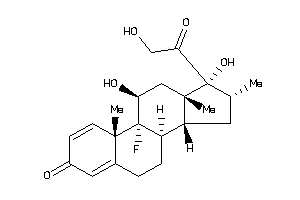
-
3978014
-
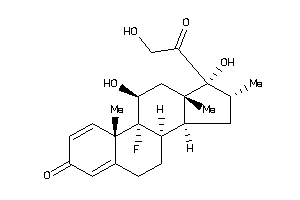
-
3978043
-
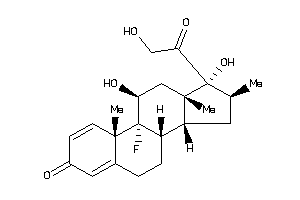
-
3978044
-
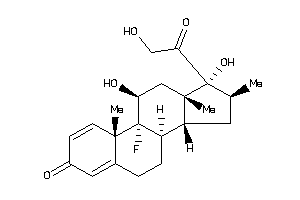
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.38 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Functional ≤ 10μM
|
|
Z50594-7-O |
Mus Musculus (cluster #7 Of 9), Other |
Other |
5 |
0.42 |
Functional ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
20 |
0.38 |
Functional ≤ 10μM
|
|
Z80178-2-O |
J774.A1 (Macrophage Cells) (cluster #2 Of 3), Other |
Other |
48 |
0.37 |
Functional ≤ 10μM
|
|
Z80301-2-O |
N9 (cluster #2 Of 2), Other |
Other |
74 |
0.36 |
Functional ≤ 10μM
|
|
Z80418-3-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #3 Of 9), Other |
Other |
860 |
0.30 |
Functional ≤ 10μM
|
|
Z80548-4-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #4 Of 5), Other |
Other |
7 |
0.41 |
Functional ≤ 10μM
|
|
Z80954-3-O |
HFF (Foreskin Fibroblasts) (cluster #3 Of 4), Other |
Other |
2 |
0.43 |
Functional ≤ 10μM
|
|
Z81247-4-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #4 Of 9), Other |
Other |
17 |
0.39 |
Functional ≤ 10μM
|
|
Z81338-2-O |
T-cells (cluster #2 Of 3), Other |
Other |
5 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
3.98 |
-16.38 |
3 |
5 |
0 |
95 |
392.467 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
10.37 |
-11.66 |
0 |
2 |
0 |
34 |
312.453 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
3.62 |
10.62 |
-10.79 |
0 |
2 |
0 |
34 |
312.453 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10S,11S,13S,14S,16R,17R)-9-fluoro-11,17-dihydroxy-10,13,16-trimethyl-3-oxo-6,7,8,11,12,14,15,
(8S,9R,10S,11S,13S,14S,16R,17R)-…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80110-2-O |
CV-1 (Kidney Cells) (cluster #2 Of 2), Other |
Other |
6 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
6.36 |
-52.9 |
2 |
5 |
-1 |
98 |
377.432 |
1 |
↓
|
|
|
|
Analogs
-
34633676
-
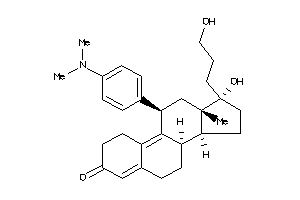
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
8.94 |
-11.75 |
2 |
4 |
0 |
61 |
449.635 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
3830334
-
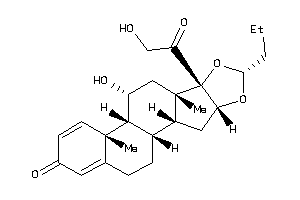
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.38 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.36 |
Functional ≤ 10μM |
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
1 |
0.41 |
Functional ≤ 10μM |
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
3162 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
6.66 |
-12.84 |
2 |
6 |
0 |
93 |
430.541 |
4 |
↓
|
|
|
|
|
|
|
|