|
|
Analogs
-
19850502
-
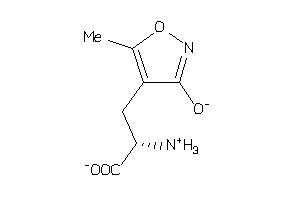
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
364 |
0.69 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
364 |
0.69 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
40 |
0.80 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
40 |
0.80 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Binding ≤ 10μM
|
|
GRIK1-1-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
740 |
0.66 |
Binding ≤ 10μM
|
|
GRIK2-3-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.85 |
Binding ≤ 10μM
|
|
GRIK3-3-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.85 |
Binding ≤ 10μM
|
|
GRIK4-1-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.85 |
Binding ≤ 10μM
|
|
GRIK5-1-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.85 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3310 |
0.59 |
Functional ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3310 |
0.59 |
Functional ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.59 |
Functional ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.59 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
800 |
0.66 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
4800 |
0.57 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.69 |
-1.91 |
-52.1 |
3 |
6 |
-1 |
117 |
185.159 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.69 |
-2.52 |
-37.61 |
4 |
6 |
0 |
114 |
186.167 |
3 |
↓
|
|
|
|
Analogs
-
19850499
-
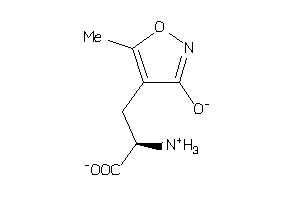
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
22 |
0.82 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
364 |
0.69 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.84 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
364 |
0.69 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
21 |
0.83 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
40 |
0.80 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
40 |
0.80 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
40 |
0.80 |
Binding ≤ 10μM
|
|
GRIK1-1-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
740 |
0.66 |
Binding ≤ 10μM
|
|
GRIK1-1-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1150 |
0.64 |
Binding ≤ 10μM
|
|
GRIK2-3-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.85 |
Binding ≤ 10μM
|
|
GRIK3-3-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.85 |
Binding ≤ 10μM
|
|
GRIK4-1-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.85 |
Binding ≤ 10μM
|
|
GRIK5-1-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.85 |
Binding ≤ 10μM
|
|
GRIK5-1-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2144 |
0.61 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3310 |
0.59 |
Functional ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.59 |
Functional ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3310 |
0.59 |
Functional ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3500 |
0.59 |
Functional ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.59 |
Functional ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.59 |
Functional ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.59 |
Functional ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.59 |
Functional ≤ 10μM
|
|
GRIK3-1-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.58 |
Functional ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
3500 |
0.59 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
800 |
0.66 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
6700 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.69 |
-1.72 |
-76.52 |
3 |
6 |
-1 |
117 |
185.159 |
3 |
↓
|
|
|
|
Analogs
-
19850510
-
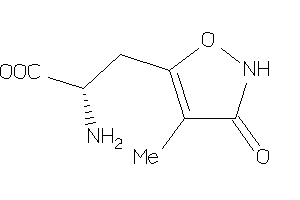
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.68 |
-0.55 |
-39.54 |
3 |
6 |
-1 |
112 |
185.159 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.23 |
-2.22 |
-94.17 |
2 |
6 |
-2 |
115 |
184.151 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.23 |
-3.13 |
-40.73 |
4 |
6 |
0 |
114 |
186.167 |
3 |
↓
|
|
|
|
Analogs
-
19850508
-
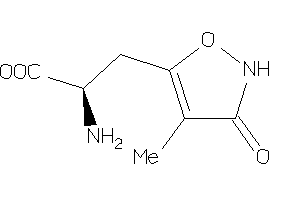
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.68 |
-0.42 |
-41.3 |
3 |
6 |
-1 |
112 |
185.159 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.23 |
-2.09 |
-96.11 |
2 |
6 |
-2 |
115 |
184.151 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.23 |
-3.06 |
-44.01 |
4 |
6 |
0 |
114 |
186.167 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2100 |
0.50 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2780 |
0.49 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.50 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2100 |
0.50 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2100 |
0.50 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7900 |
0.45 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
2100 |
0.50 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7600 |
0.45 |
Binding ≤ 10μM
|
|
GRIK1-1-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.76 |
Binding ≤ 10μM
|
|
GRIK3-3-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2530 |
0.49 |
Binding ≤ 10μM
|
|
GRIK5-1-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
480 |
0.55 |
Binding ≤ 10μM
|
|
GRIK1-1-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-0.38 |
-49.09 |
3 |
6 |
-1 |
117 |
227.24 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.97 |
-0.69 |
-105.71 |
2 |
6 |
-2 |
115 |
226.232 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.42 |
1.24 |
-28.43 |
4 |
6 |
0 |
114 |
228.248 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.32 |
Binding ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.32 |
Binding ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.32 |
Binding ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
9000 |
0.32 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50501-1-O |
Gallus Gallus (cluster #1 Of 3), Other |
Other |
9500 |
0.32 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
9900 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
5.48 |
-12.21 |
2 |
5 |
0 |
69 |
293.326 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
3.69 |
-62.71 |
2 |
11 |
-1 |
155 |
515.568 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.16 |
6.1 |
-101.19 |
3 |
11 |
0 |
157 |
516.576 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.16 |
5.54 |
-97.21 |
3 |
11 |
0 |
157 |
516.576 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
3.68 |
-62.27 |
2 |
11 |
-1 |
155 |
515.568 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.16 |
6.1 |
-99.52 |
3 |
11 |
0 |
157 |
516.576 |
8 |
↓
|
|
|
|
Analogs
-
26378421
-
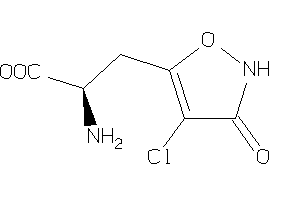
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
130 |
0.74 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
370 |
0.69 |
Binding ≤ 10μM
|
|
GRIK1-1-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6800 |
0.56 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4700 |
0.57 |
Functional ≤ 10μM
|
|
GRIA2-2-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.45 |
-0.69 |
-40.56 |
3 |
6 |
-1 |
112 |
205.577 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.00 |
-2.71 |
-32.85 |
4 |
6 |
0 |
114 |
206.585 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.00 |
-2.24 |
-90.61 |
2 |
6 |
-2 |
115 |
204.569 |
3 |
↓
|
|
|
|
Analogs
-
26378419
-
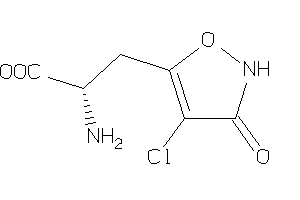
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
130 |
0.74 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
370 |
0.69 |
Binding ≤ 10μM
|
|
GRIK1-1-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6800 |
0.56 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4700 |
0.57 |
Functional ≤ 10μM
|
|
GRIA2-2-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.45 |
-0.72 |
-39.44 |
3 |
6 |
-1 |
112 |
205.577 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.00 |
-2.36 |
-92.35 |
2 |
6 |
-2 |
115 |
204.569 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.00 |
-2.78 |
-33.52 |
4 |
6 |
0 |
114 |
206.585 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
170 |
0.73 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
340 |
0.70 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
250 |
0.71 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
340 |
0.70 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
340 |
0.70 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
340 |
0.70 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM
|
|
GRIK1-1-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Binding ≤ 10μM
|
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.72 |
Binding ≤ 10μM
|
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.72 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.72 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.72 |
Binding ≤ 10μM
|
|
GRM7-1-E |
Metabotropic Glutamate Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.72 |
Binding ≤ 10μM
|
|
GRM7-1-E |
Metabotropic Glutamate Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.72 |
Binding ≤ 10μM
|
|
GRM8-2-E |
Metabotropic Glutamate Receptor 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.72 |
Binding ≤ 10μM
|
|
GRM8-2-E |
Metabotropic Glutamate Receptor 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.72 |
Binding ≤ 10μM
|
|
Q80T35-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.72 |
Binding ≤ 10μM
|
|
Q80T35-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.72 |
Binding ≤ 10μM
|
|
Q80T43-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.72 |
Binding ≤ 10μM
|
|
Q80T43-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.72 |
Binding ≤ 10μM
|
|
Q80T47-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.72 |
Binding ≤ 10μM
|
|
Q80T47-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.72 |
Binding ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5400 |
0.57 |
Functional ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Functional ≤ 10μM
|
|
GRIA4-2-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
200 |
0.72 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
220 |
0.72 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.32 |
-0.57 |
-40.2 |
3 |
6 |
-1 |
112 |
250.028 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.86 |
-2.1 |
-91.82 |
2 |
6 |
-2 |
115 |
249.02 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.32 |
-0.24 |
-33.53 |
4 |
6 |
0 |
114 |
251.036 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2100 |
0.50 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2100 |
0.50 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2100 |
0.50 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
2100 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-0.34 |
-48.46 |
3 |
6 |
-1 |
117 |
227.24 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.97 |
-0.64 |
-107.42 |
2 |
6 |
-2 |
115 |
226.232 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.97 |
-0.92 |
-35.36 |
4 |
6 |
0 |
114 |
228.248 |
4 |
↓
|
|
|
|
Analogs
-
4475051
-
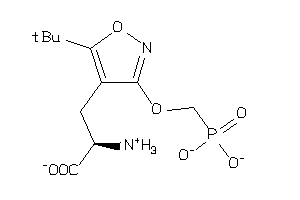
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5700 |
0.35 |
Binding ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5700 |
0.35 |
Binding ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5700 |
0.35 |
Binding ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5700 |
0.35 |
Binding ≤ 10μM
|
|
GRIK1-1-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.96 |
-2.77 |
-126.83 |
3 |
9 |
-2 |
166 |
320.238 |
7 |
↓
|
|
|
|
Analogs
-
2047684
-
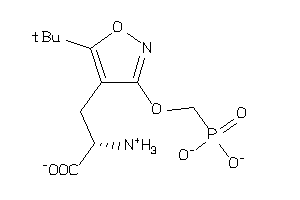
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5700 |
0.35 |
Binding ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5700 |
0.35 |
Binding ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5700 |
0.35 |
Binding ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5700 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.96 |
-2.77 |
-142.28 |
3 |
9 |
-2 |
166 |
320.238 |
7 |
↓
|
|
|
|
Analogs
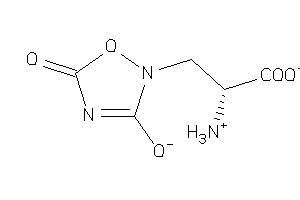
-
22066252
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FOLH1-1-E |
Glutamate Carboxypeptidase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.54 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
171 |
0.73 |
Binding ≤ 10μM
|
|
GRIK2-3-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRIK3-3-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1670 |
0.62 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRIK5-1-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.72 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRM7-1-E |
Metabotropic Glutamate Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM8-2-E |
Metabotropic Glutamate Receptor 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
Q80T35-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
Q80T43-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
Q80T47-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
XCT-1-E |
Cystine/glutamate Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.57 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1100 |
0.64 |
Functional ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
55 |
0.78 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
7300 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.04 |
-4.9 |
-43.55 |
3 |
8 |
-1 |
139 |
188.119 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.04 |
-5.21 |
-87.71 |
2 |
8 |
-2 |
137 |
187.111 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.04 |
-7.81 |
-15.85 |
4 |
8 |
0 |
132 |
189.127 |
3 |
↓
|
|
|
|
Analogs
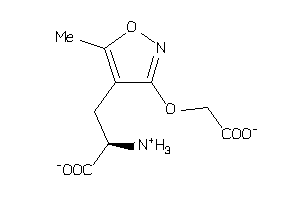
-
4293721
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.17 |
-2.64 |
-66.16 |
3 |
8 |
-1 |
143 |
243.195 |
6 |
↓
|
|
|
|
Analogs
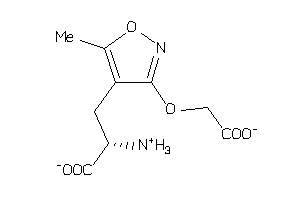
-
2562328
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.17 |
-2.41 |
-93.51 |
3 |
8 |
-1 |
143 |
243.195 |
6 |
↓
|
|