|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM |
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM |
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM |
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
-0.17 |
-9.29 |
2 |
4 |
0 |
65 |
309.041 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
37 |
0.39 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
Z104298-1-O |
Glutamate NMDA Receptor; GRIN1/GRIN2A (cluster #1 Of 2), Other |
Other |
270 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
2.81 |
-46.71 |
4 |
8 |
-1 |
138 |
453.209 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.93 |
5.23 |
-81.93 |
4 |
8 |
-1 |
146 |
453.209 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.92 |
3.94 |
-132.89 |
3 |
8 |
-2 |
141 |
452.201 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
37 |
0.39 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
Z104298-1-O |
Glutamate NMDA Receptor; GRIN1/GRIN2A (cluster #1 Of 2), Other |
Other |
270 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
3.69 |
-44.33 |
4 |
8 |
-1 |
138 |
453.209 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.93 |
3.96 |
-38.6 |
5 |
8 |
0 |
143 |
454.217 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRID1-1-E |
Glutamate Receptor Delta-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM
|
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM |
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM |
|
NMDE1-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM
|
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
3400 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.99 |
2.59 |
-8.79 |
2 |
4 |
0 |
66 |
231.038 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.90 |
0.52 |
-42.98 |
1 |
4 |
-1 |
69 |
230.03 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-2-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1640 |
0.58 |
Binding ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1640 |
0.58 |
Binding ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1640 |
0.58 |
Binding ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1640 |
0.58 |
Binding ≤ 10μM
|
|
GRID1-1-E |
Glutamate Receptor Delta-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
130 |
0.69 |
Binding ≤ 10μM
|
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
130 |
0.69 |
Binding ≤ 10μM |
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
130 |
0.69 |
Binding ≤ 10μM |
|
NMDE1-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
130 |
0.69 |
Binding ≤ 10μM
|
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
130 |
0.69 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
130 |
0.69 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
130 |
0.69 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
130 |
0.69 |
Binding ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
3100 |
0.55 |
Binding ≤ 10μM
|
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
3981 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.99 |
2.46 |
-9.86 |
2 |
4 |
0 |
66 |
231.038 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.86 |
1.7 |
-36.44 |
1 |
4 |
1 |
61 |
230.03 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.90 |
0.31 |
-40.97 |
1 |
4 |
-1 |
69 |
230.03 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
F16P1-1-E |
Fructose-1,6-bisphosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.41 |
Binding ≤ 10μM
|
|
F16P2-1-E |
Fructose-1,6-bisphosphatase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.41 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2700 |
0.41 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
70 |
0.53 |
Binding ≤ 10μM |
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
50 |
0.54 |
Binding ≤ 10μM |
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
5000 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
7.85 |
-111.23 |
1 |
5 |
-2 |
96 |
300.097 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-2-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
360 |
0.50 |
Binding ≤ 10μM
|
|
GRIA2-2-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
360 |
0.50 |
Binding ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
360 |
0.50 |
Binding ≤ 10μM
|
|
GRIA4-2-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
360 |
0.50 |
Binding ≤ 10μM
|
|
GRIK1-2-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.43 |
Binding ≤ 10μM |
|
GRIK2-1-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.43 |
Binding ≤ 10μM |
|
GRIK3-1-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.43 |
Binding ≤ 10μM |
|
GRIK4-1-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.43 |
Binding ≤ 10μM |
|
GRIK5-1-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
740 |
0.48 |
Binding ≤ 10μM
|
|
NMD3A-4-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.50 |
Binding ≤ 10μM
|
|
NMD3B-4-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.50 |
Binding ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
840 |
0.47 |
Binding ≤ 10μM |
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
840 |
0.47 |
Binding ≤ 10μM |
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
840 |
0.47 |
Binding ≤ 10μM |
|
NMDE4-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.50 |
Binding ≤ 10μM
|
|
NMDZ1-1-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
840 |
0.47 |
Binding ≤ 10μM |
|
Z104302-5-O |
Glutamate NMDA Receptor (cluster #5 Of 7), Other |
Other |
4500 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.46 |
1.23 |
-35.53 |
1 |
10 |
-1 |
160 |
251.134 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
0.42 |
3.67 |
-137.39 |
2 |
10 |
2 |
154 |
252.142 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.42 |
2.62 |
-51.15 |
1 |
10 |
1 |
153 |
251.134 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
NMDZ1-1-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
400 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
6.77 |
-40.47 |
1 |
3 |
-1 |
56 |
238.675 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.09 |
5.5 |
-102.71 |
0 |
3 |
-2 |
53 |
237.667 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
|
Z104297-2-O |
Glutamate NMDA Receptor; GRIN1/GRIN2B (cluster #2 Of 3), Other |
Other |
6 |
0.40 |
Binding ≤ 10μM |
|
Z104297-1-O |
Glutamate NMDA Receptor; GRIN1/GRIN2B (cluster #1 Of 1), Other |
Other |
70 |
0.35 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
3.91 |
-53.36 |
1 |
7 |
-1 |
98 |
396.398 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.42 |
5.62 |
-21.27 |
2 |
7 |
0 |
95 |
397.406 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRID1-1-E |
Glutamate Receptor Delta-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.62 |
Binding ≤ 10μM
|
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
1800 |
0.62 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
1800 |
0.62 |
Binding ≤ 10μM
|
|
NMDE1-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1800 |
0.62 |
Binding ≤ 10μM
|
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1800 |
0.62 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.62 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
1800 |
0.62 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1800 |
0.62 |
Binding ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
8400 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
-7.16 |
-8.4 |
2 |
4 |
0 |
66 |
196.593 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRID1-1-E |
Glutamate Receptor Delta-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.55 |
Binding ≤ 10μM
|
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
3300 |
0.55 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
3300 |
0.55 |
Binding ≤ 10μM
|
|
NMDE1-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
3300 |
0.55 |
Binding ≤ 10μM
|
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
3300 |
0.55 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3300 |
0.55 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
3300 |
0.55 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
3300 |
0.55 |
Binding ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
2300 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
2.7 |
-10.8 |
2 |
4 |
0 |
66 |
190.202 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRID1-1-E |
Glutamate Receptor Delta-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9800 |
0.58 |
Binding ≤ 10μM
|
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
9800 |
0.58 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
9800 |
0.58 |
Binding ≤ 10μM
|
|
NMDE1-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
9800 |
0.58 |
Binding ≤ 10μM
|
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
9800 |
0.58 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9800 |
0.58 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
9800 |
0.58 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
9800 |
0.58 |
Binding ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
9800 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.27 |
1.48 |
-11.66 |
2 |
4 |
0 |
66 |
162.148 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRID1-1-E |
Glutamate Receptor Delta-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.57 |
Binding ≤ 10μM
|
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
2100 |
0.57 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
2100 |
0.57 |
Binding ≤ 10μM
|
|
NMDE1-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
2100 |
0.57 |
Binding ≤ 10μM
|
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
2100 |
0.57 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2100 |
0.57 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
2100 |
0.57 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
2100 |
0.57 |
Binding ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
2100 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
2.94 |
-10.71 |
2 |
4 |
0 |
66 |
190.202 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRID1-1-E |
Glutamate Receptor Delta-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.59 |
Binding ≤ 10μM
|
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
1400 |
0.59 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
1400 |
0.59 |
Binding ≤ 10μM
|
|
NMDE1-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1400 |
0.59 |
Binding ≤ 10μM
|
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1400 |
0.59 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.59 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
1400 |
0.59 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1400 |
0.59 |
Binding ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
1400 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.51 |
1.01 |
-11.51 |
2 |
5 |
0 |
83 |
190.158 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.39 |
0.03 |
-122.56 |
0 |
5 |
-2 |
88 |
188.142 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.39 |
0.42 |
-128.47 |
0 |
5 |
-2 |
88 |
188.142 |
0 |
↓
|
|
|
|
Analogs
-
34354758
-
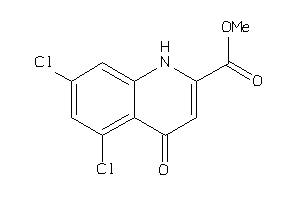
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLRA1-1-E |
Glycine Receptor Subunit Alpha-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GLRA2-1-E |
Glycine Receptor Subunit Alpha-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GLRA3-1-E |
Glycine Receptor Alpha-3 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GLRB-1-E |
Glycine Receptor Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GRIA1-2-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE1-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMD3B-1-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
90 |
0.62 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
5.87 |
-38.83 |
1 |
4 |
-1 |
73 |
257.052 |
1 |
↓
|
|