|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNCG-1-E |
Phosphodiesterase 6H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.31 |
Binding ≤ 10μM
|
|
CNRG-1-E |
Phosphodiesterase 6G (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.31 |
Binding ≤ 10μM
|
|
E1BN64-1-E |
Phosphodiesterase 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9200 |
0.21 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5480 |
0.22 |
Binding ≤ 10μM
|
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.23 |
Binding ≤ 10μM
|
|
PDE10-1-E |
Phosphodiesterase 10A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.23 |
Binding ≤ 10μM
|
|
PDE11-1-E |
Phosphodiesterase 11A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.23 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.27 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.28 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.28 |
Binding ≤ 10μM
|
|
PDE3A-1-E |
Phosphodiesterase 3A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.22 |
Binding ≤ 10μM
|
|
PDE3B-1-E |
Phosphodiesterase 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.22 |
Binding ≤ 10μM
|
|
PDE4A-3-E |
Phosphodiesterase 4A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4600 |
0.23 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9600 |
0.21 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9600 |
0.21 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9600 |
0.21 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.35 |
Binding ≤ 10μM
|
|
PDE6A-1-E |
Rod CGMP-specific 3',5'-cyclic Phosphodiesterase Subunit Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.31 |
Binding ≤ 10μM
|
|
PDE6B-1-E |
Phosphodiesterase 6B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.31 |
Binding ≤ 10μM
|
|
PDE6C-1-E |
Phosphodiesterase 6C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.31 |
Binding ≤ 10μM
|
|
PDE6D-1-E |
Phosphodiesterase 6D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
74 |
0.30 |
Binding ≤ 10μM
|
|
PDE9A-1-E |
Phosphodiesterase 9A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.23 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.34 |
Binding ≤ 10μM
|
|
Q9EPR9-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.28 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.34 |
Functional ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.33 |
Functional ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
38 |
0.31 |
Functional ≤ 10μM
|
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 8), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
4.42 |
-15.04 |
1 |
10 |
0 |
113 |
474.587 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.51 |
4.17 |
-20.18 |
1 |
10 |
0 |
113 |
474.587 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.97 |
2.87 |
-54.77 |
0 |
10 |
-1 |
117 |
473.579 |
7 |
↓
|
|
|
|
Analogs
-
8204637
-
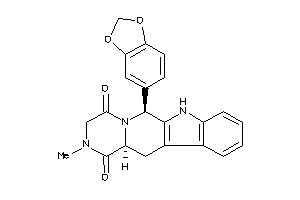
-
8204642
-
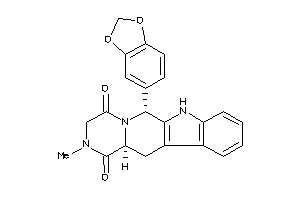
-
538404
-
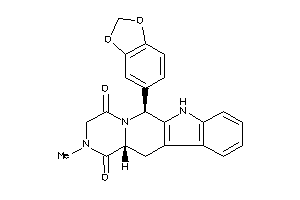
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNCG-1-E |
Phosphodiesterase 6H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.29 |
Binding ≤ 10μM
|
|
CNRG-1-E |
Phosphodiesterase 6G (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.29 |
Binding ≤ 10μM
|
|
PDE11-1-E |
Phosphodiesterase 11A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
PDE6A-1-E |
Rod CGMP-specific 3',5'-cyclic Phosphodiesterase Subunit Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.29 |
Binding ≤ 10μM
|
|
PDE6B-1-E |
Phosphodiesterase 6B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.29 |
Binding ≤ 10μM
|
|
PDE6C-1-E |
Phosphodiesterase 6C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.29 |
Binding ≤ 10μM
|
|
PDE6D-1-E |
Phosphodiesterase 6D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5100 |
0.26 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
500 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.36 |
6.81 |
-13.33 |
1 |
7 |
0 |
75 |
389.411 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
6.48 |
-10.31 |
0 |
5 |
0 |
52 |
270.336 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
13.32 |
-49.29 |
1 |
10 |
-1 |
121 |
452.539 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.84 |
13.83 |
-64.77 |
2 |
10 |
0 |
122 |
453.547 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
8.28 |
-9.75 |
2 |
4 |
0 |
54 |
385.261 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5600 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
8.55 |
-6.96 |
2 |
4 |
0 |
54 |
385.261 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
9.58 |
-10.08 |
1 |
3 |
0 |
38 |
263.344 |
4 |
↓
|
|
|
|
Analogs
-
9059078
-
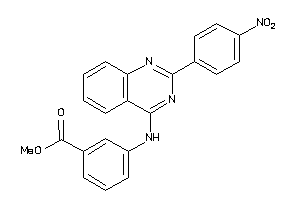
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
1.64 |
-12.49 |
1 |
5 |
0 |
64 |
355.397 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.93 |
10.93 |
-9.51 |
1 |
3 |
0 |
38 |
317.417 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.93 |
11.28 |
-24.03 |
2 |
3 |
1 |
39 |
318.425 |
4 |
↓
|
|
|
|
Analogs
-
25150455
-
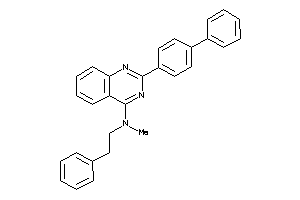
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.44 |
-0.57 |
-11.42 |
1 |
3 |
0 |
37 |
325.415 |
5 |
↓
|
|
|
|
Analogs
-
53160544
-
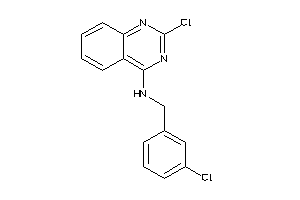
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
-1.62 |
-8.28 |
1 |
3 |
0 |
37 |
269.735 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.91 |
0.66 |
-13.2 |
1 |
4 |
0 |
50 |
287.322 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
-1.52 |
-9.48 |
1 |
4 |
0 |
50 |
312.376 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
10.24 |
-10.49 |
1 |
4 |
0 |
51 |
298.349 |
3 |
↓
|
|
|
|
Analogs
-
25142577
-
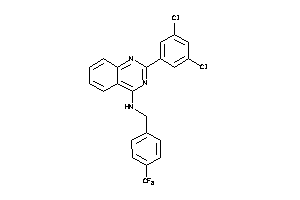
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.69 |
-0.85 |
-9.87 |
1 |
3 |
0 |
37 |
345.833 |
4 |
↓
|
|
|
|
Analogs
-
25142577
-

-
25151836
-
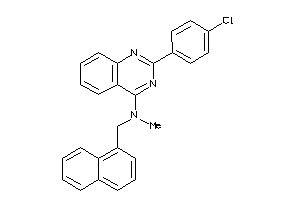
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
740 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.71 |
-0.85 |
-9.62 |
1 |
3 |
0 |
37 |
345.833 |
4 |
↓
|
|
|
|
Analogs
-
9087670
-
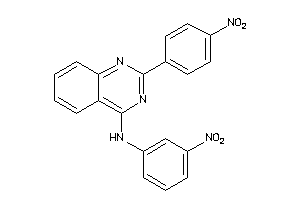
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
420 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.70 |
1.02 |
-12.04 |
1 |
6 |
0 |
83 |
342.358 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.72 |
-4.06 |
-9.24 |
1 |
5 |
0 |
50 |
320.396 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
10.97 |
-9.64 |
1 |
3 |
0 |
38 |
289.382 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.03 |
11.32 |
-22.91 |
2 |
3 |
1 |
39 |
290.39 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
10.03 |
-9.85 |
1 |
4 |
0 |
51 |
312.376 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.96 |
10.38 |
-26.17 |
2 |
4 |
1 |
52 |
313.384 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
860 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.94 |
1.63 |
-12.94 |
1 |
5 |
0 |
64 |
355.397 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 43 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNCG-1-E |
Phosphodiesterase 6H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.33 |
Binding ≤ 10μM
|
|
CNRG-1-E |
Phosphodiesterase 6G (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.33 |
Binding ≤ 10μM
|
|
PDE10-1-E |
Phosphodiesterase 10A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
790 |
0.25 |
Binding ≤ 10μM
|
|
PDE11-1-E |
Phosphodiesterase 11A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
130 |
0.28 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.27 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.27 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.27 |
Binding ≤ 10μM
|
|
PDE3A-1-E |
Phosphodiesterase 3A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.23 |
Binding ≤ 10μM
|
|
PDE3B-1-E |
Phosphodiesterase 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.23 |
Binding ≤ 10μM
|
|
PDE4A-3-E |
Phosphodiesterase 4A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4500 |
0.22 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.22 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.22 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.22 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM
|
|
PDE6A-1-E |
Rod CGMP-specific 3',5'-cyclic Phosphodiesterase Subunit Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.33 |
Binding ≤ 10μM
|
|
PDE6B-1-E |
Phosphodiesterase 6B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.33 |
Binding ≤ 10μM
|
|
PDE6C-1-E |
Phosphodiesterase 6C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.33 |
Binding ≤ 10μM
|
|
PDE6D-1-E |
Phosphodiesterase 6D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.33 |
Binding ≤ 10μM
|
|
PDE9A-1-E |
Phosphodiesterase 9A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
6.46 |
-15.5 |
1 |
10 |
0 |
113 |
488.614 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.01 |
4.67 |
-56.65 |
0 |
10 |
-1 |
116 |
487.606 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.56 |
8.68 |
-54.65 |
2 |
10 |
1 |
114 |
489.622 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.84 |
8.96 |
-6.18 |
2 |
4 |
0 |
54 |
375.255 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.84 |
8.7 |
-9.23 |
2 |
4 |
0 |
54 |
375.255 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
3.14 |
-18.02 |
2 |
11 |
0 |
133 |
504.613 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.01 |
1.36 |
-59.12 |
1 |
11 |
-1 |
136 |
503.605 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.49 |
-1.75 |
-52.85 |
2 |
5 |
-1 |
81 |
369.828 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.37 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.37 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.37 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
5.81 |
-9.95 |
3 |
5 |
0 |
73 |
318.427 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.55 |
6.21 |
-29.79 |
4 |
5 |
1 |
74 |
319.435 |
4 |
↓
|
|
|
|
Analogs
-
20348532
-
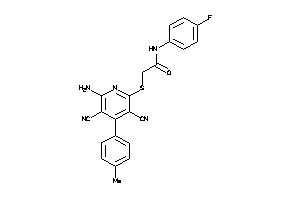
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5700 |
0.25 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5700 |
0.25 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5700 |
0.25 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
8.91 |
-19.99 |
3 |
6 |
0 |
116 |
403.442 |
5 |
↓
|
|
|
|
Analogs
-
39853015
-
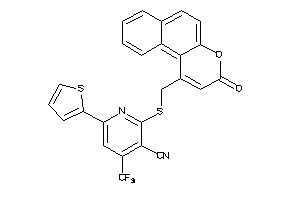
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.29 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.29 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.29 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.35 |
2.75 |
-54.16 |
0 |
4 |
-1 |
76 |
419.429 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.35 |
2.97 |
-77.86 |
1 |
4 |
0 |
78 |
420.437 |
6 |
↓
|
|
|
|
Analogs
-
4550964
-
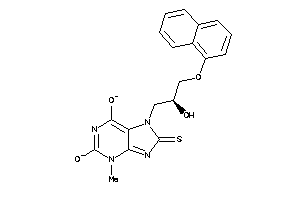
-
5447786
-
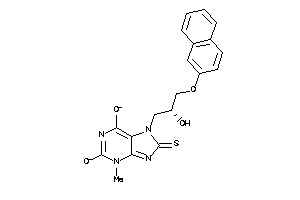
-
627151
-
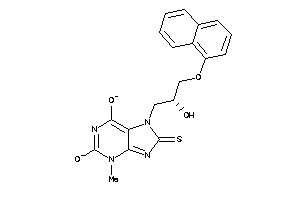
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
2.98 |
-98.97 |
1 |
8 |
-2 |
111 |
396.428 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.25 |
6.3 |
-16.65 |
3 |
8 |
0 |
105 |
398.444 |
5 |
↓
|
|
|
|
Analogs
-
627151
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
2.46 |
-101.02 |
1 |
8 |
-2 |
111 |
396.428 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.25 |
6.81 |
-17.05 |
3 |
8 |
0 |
105 |
398.444 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3700 |
0.27 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3700 |
0.27 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3700 |
0.27 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
-1.65 |
-54.88 |
1 |
6 |
-1 |
99 |
391.428 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6300 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
5.39 |
-17.47 |
3 |
6 |
0 |
102 |
423.519 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.46 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.45 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9100 |
0.44 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
800 |
0.53 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.48 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.48 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.48 |
Binding ≤ 10μM
|
|
PDE3A-1-E |
Phosphodiesterase 3A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE3B-1-E |
Phosphodiesterase 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE4A-3-E |
Phosphodiesterase 4A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE4B-2-E |
Phosphodiesterase 4B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE4C-2-E |
Phosphodiesterase 4C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE4D-2-E |
Phosphodiesterase 4D (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.45 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3020 |
0.48 |
Binding ≤ 10μM
|
|
PDE7A-1-E |
Phosphodiesterase 7A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.47 |
Binding ≤ 10μM
|
|
PDE7B-1-E |
Phosphodiesterase 7B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.47 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.50 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.50 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.50 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.50 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.45 |
Functional ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.46 |
Functional ≤ 10μM
|
|
PDE1B-2-E |
Phosphodiesterase 1B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.61 |
Functional ≤ 10μM
|
|
PDE1C-2-E |
Phosphodiesterase 1C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.61 |
Functional ≤ 10μM
|
|
Q9EPR9-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.61 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 5), Other |
Other |
800 |
0.53 |
Binding ≤ 10μM
|
|
Z50588-1-O |
Canis Familiaris (cluster #1 Of 7), Other |
Other |
5500 |
0.46 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3500 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
5.25 |
-10.93 |
1 |
6 |
0 |
73 |
222.248 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNCG-1-E |
Phosphodiesterase 6H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
CNRG-1-E |
Phosphodiesterase 6G (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9400 |
0.35 |
Binding ≤ 10μM |
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9400 |
0.35 |
Binding ≤ 10μM |
|
PDE4D-2-E |
Phosphodiesterase 4D (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8760 |
0.35 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.44 |
Binding ≤ 10μM
|
|
PDE6A-1-E |
Rod CGMP-specific 3',5'-cyclic Phosphodiesterase Subunit Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
PDE6B-1-E |
Phosphodiesterase 6B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
PDE6C-1-E |
Phosphodiesterase 6C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
PDE6D-1-E |
Phosphodiesterase 6D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
|
Q9EPR9-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9400 |
0.35 |
Binding ≤ 10μM |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.35 |
Functional ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
4750 |
0.37 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
9000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
2.82 |
-126.49 |
0 |
7 |
-2 |
98 |
269.264 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.59 |
2.52 |
-46.12 |
1 |
7 |
-1 |
100 |
270.272 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.59 |
2.48 |
-50.96 |
1 |
7 |
-1 |
100 |
270.272 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.39 |
Binding ≤ 10μM |
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.39 |
Binding ≤ 10μM |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
82 |
0.45 |
Binding ≤ 10μM |
|
Q9EPR9-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.39 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.72 |
4.08 |
-51.84 |
1 |
6 |
-1 |
87 |
297.338 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.26 |
5.7 |
-9.97 |
2 |
6 |
0 |
84 |
298.346 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.26 |
6.08 |
-14.02 |
2 |
6 |
0 |
84 |
298.346 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.42 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
1.42 |
-56.8 |
2 |
6 |
-1 |
98 |
269.284 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.61 |
2.69 |
-9.7 |
3 |
6 |
0 |
95 |
270.292 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.61 |
3.02 |
-13.64 |
3 |
6 |
0 |
95 |
270.292 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3900 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.54 |
11.5 |
-11.15 |
1 |
3 |
0 |
38 |
303.409 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.54 |
11.82 |
-25.37 |
2 |
3 |
1 |
39 |
304.417 |
3 |
↓
|
|
|
|
Analogs
-
5744976
-
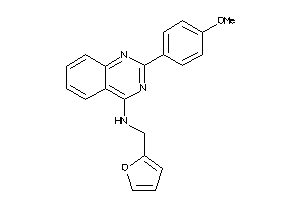
-
25145846
-
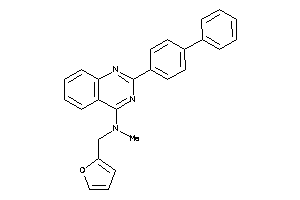
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.29 |
9.95 |
-10.36 |
1 |
4 |
0 |
51 |
301.349 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.29 |
10.3 |
-24.86 |
2 |
4 |
1 |
52 |
302.357 |
4 |
↓
|
|
|
|
Analogs
-
25142577
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.66 |
-0.88 |
-10.22 |
1 |
3 |
0 |
37 |
345.833 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.40 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.40 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.40 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
0.6 |
-11.93 |
1 |
6 |
0 |
72 |
312.373 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3250 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.09 |
2.54 |
-21.58 |
3 |
6 |
0 |
91 |
277.305 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.09 |
3.76 |
-62.51 |
2 |
6 |
-1 |
94 |
276.297 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.42 |
Binding ≤ 10μM
|
|
PDE6C-1-E |
Phosphodiesterase 6C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
7.49 |
-15.28 |
1 |
7 |
0 |
90 |
354.41 |
6 |
↓
|
|
|
|
Analogs
-
38155979
-
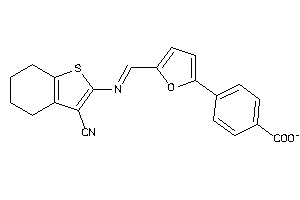
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.20 |
1.55 |
-61.93 |
0 |
5 |
-1 |
89 |
375.429 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8600 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.80 |
10.03 |
-11.05 |
1 |
4 |
0 |
51 |
312.376 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.80 |
10.38 |
-26.69 |
2 |
4 |
1 |
52 |
313.384 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.80 |
10.84 |
-77.23 |
3 |
4 |
2 |
53 |
314.392 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
81 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
3.19 |
-46.77 |
1 |
5 |
-1 |
78 |
253.285 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.88 |
3.99 |
-20 |
2 |
5 |
0 |
74 |
254.293 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.25 |
Binding ≤ 10μM |
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.25 |
Binding ≤ 10μM |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.31 |
Binding ≤ 10μM |
|
Q9EPR9-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.25 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
1.91 |
-17.31 |
2 |
11 |
0 |
134 |
504.613 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.34 |
0.36 |
-56.53 |
1 |
11 |
-1 |
137 |
503.605 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.88 |
1.66 |
-22.13 |
2 |
11 |
0 |
134 |
504.613 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
790 |
0.37 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
790 |
0.37 |
Binding ≤ 10μM |
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
790 |
0.37 |
Binding ≤ 10μM |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.45 |
Binding ≤ 10μM
|
|
Q9EPR9-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
790 |
0.37 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
6.99 |
-10.77 |
1 |
6 |
0 |
73 |
312.373 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.79 |
5.44 |
-53.1 |
0 |
6 |
-1 |
76 |
311.365 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
360 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
9.15 |
-9.8 |
1 |
4 |
0 |
51 |
318.405 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.86 |
9.5 |
-26.35 |
2 |
4 |
1 |
52 |
319.413 |
4 |
↓
|
|