|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3990 |
0.25 |
Binding ≤ 10μM
|
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
2.55 |
-11.17 |
4 |
6 |
0 |
99 |
434.554 |
7 |
↓
|
|
|
|
Analogs
-
4083068
-
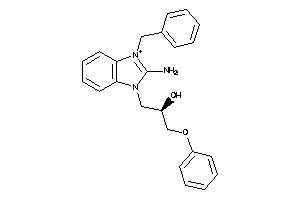
-
4116227
-
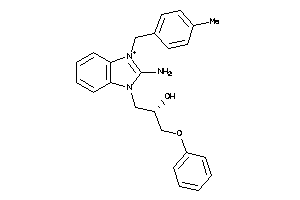
-
1302176
-
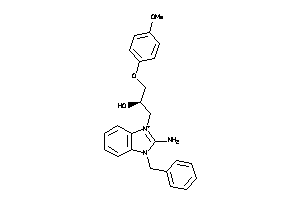
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.26 |
12.44 |
-35.42 |
3 |
5 |
1 |
64 |
374.464 |
7 |
↓
|
|
|
|
Analogs
-
4116227
-

-
1302176
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.26 |
12.51 |
-35.5 |
3 |
5 |
1 |
64 |
374.464 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9QXI6-1-E |
SGLT1 Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.25 |
Binding ≤ 10μM
|
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.23 |
Binding ≤ 10μM
|
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
13.76 |
-9.49 |
0 |
5 |
0 |
51 |
484.956 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3850 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.28 |
11.6 |
-29.7 |
3 |
4 |
1 |
55 |
324.448 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3850 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.28 |
11.68 |
-29.7 |
3 |
4 |
1 |
55 |
324.448 |
5 |
↓
|
|
|
|
Analogs
-
4164735
-
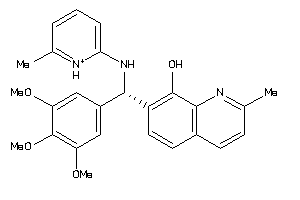
-
15015904
-
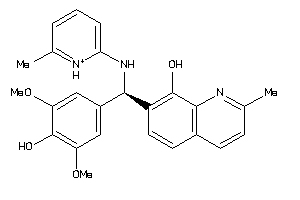
-
15015905
-
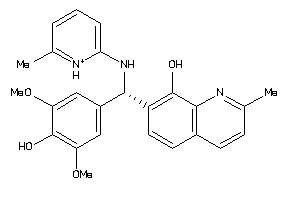
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5920 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
-1.14 |
-41.56 |
3 |
7 |
1 |
86 |
446.527 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.50 |
-0.99 |
-85.25 |
4 |
7 |
2 |
88 |
447.535 |
7 |
↓
|
|
|
|
Analogs
-
15015904
-

-
15015905
-

-
4164730
-
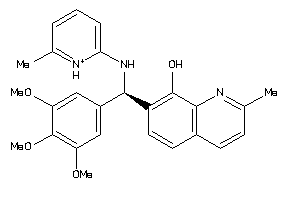
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5920 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
-1.13 |
-42.03 |
3 |
7 |
1 |
86 |
446.527 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.50 |
-0.98 |
-84.61 |
4 |
7 |
2 |
88 |
447.535 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8980 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
-5.54 |
-15.44 |
5 |
6 |
0 |
88 |
398.513 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-2-E |
Beta-secretase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5200 |
0.24 |
Binding ≤ 10μM
|
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4100 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.12 |
-2.85 |
-10.58 |
4 |
6 |
0 |
107 |
424.493 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-2-E |
Beta-secretase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7200 |
0.22 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4600 |
0.23 |
Binding ≤ 10μM
|
|
PGH1-5-E |
Cyclooxygenase-1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
600 |
0.27 |
Binding ≤ 10μM
|
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.19 |
6.87 |
-13.88 |
3 |
6 |
0 |
96 |
438.52 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-2-E |
Beta-secretase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7200 |
0.22 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4600 |
0.23 |
Binding ≤ 10μM
|
|
PGH1-5-E |
Cyclooxygenase-1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
600 |
0.27 |
Binding ≤ 10μM
|
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.19 |
6.69 |
-13.48 |
3 |
6 |
0 |
96 |
438.52 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
36520252
-
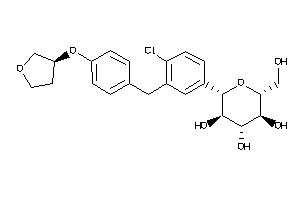
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
890 |
0.30 |
Binding ≤ 10μM
|
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
SC5A4-1-E |
Low Affinity Sodium-glucose Cotransporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
SC5AB-1-E |
Sodium/myo-inositol Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
-8.97 |
-10.83 |
4 |
6 |
0 |
99 |
408.878 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1960 |
0.27 |
Binding ≤ 10μM
|
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Binding ≤ 10μM
|
|
SC5A4-1-E |
Low Affinity Sodium-glucose Cotransporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
-1.04 |
-11.37 |
4 |
7 |
0 |
109 |
436.888 |
6 |
↓
|
|