|
|
Analogs
-
8214681
-
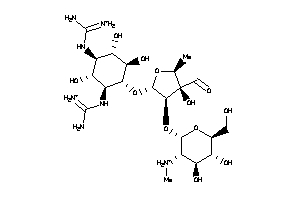
-
8214760
-
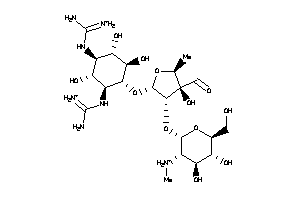
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TARB1-1-E |
TAR RNA Binding Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9500 |
0.18 |
Binding ≤ 10μM |
|
TRBP2-1-E |
TAR RNA Binding Protein 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9500 |
0.18 |
Binding ≤ 10μM |
|
Z50212-7-O |
Escherichia Coli (cluster #7 Of 7), Other |
Other |
1050 |
0.21 |
Functional ≤ 10μM
|
|
Z50380-2-O |
Mycobacterium Smegmatis (cluster #2 Of 4), Other |
Other |
500 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.28 |
-15.28 |
-142.69 |
19 |
19 |
3 |
339 |
584.604 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.28 |
-16.72 |
-90.55 |
18 |
19 |
2 |
335 |
583.596 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-2-O |
Mycobacterium Smegmatis (cluster #2 Of 4), Other |
Other |
2120 |
0.17 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-2-O |
Mycobacterium Smegmatis (cluster #2 Of 4), Other |
Other |
2120 |
0.17 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
Analogs
-
16248873
-
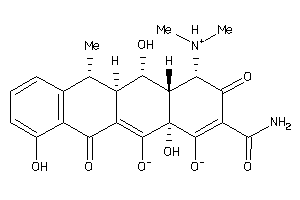
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.43 |
0.98 |
-89.55 |
6 |
10 |
-1 |
188 |
443.432 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-0.56 |
-4.34 |
-53.29 |
6 |
10 |
-1 |
185 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.61 |
-2.24 |
-69.99 |
6 |
10 |
-1 |
184 |
443.432 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
0.59 |
-95.46 |
6 |
10 |
-1 |
188 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
-1.58 |
-146.85 |
5 |
10 |
-2 |
187 |
442.424 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.24 |
-0.62 |
-247.57 |
4 |
10 |
-3 |
190 |
441.416 |
2 |
↓
|
|
|
|
Analogs
-
34013998
-
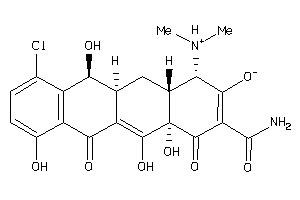
-
13719774
-
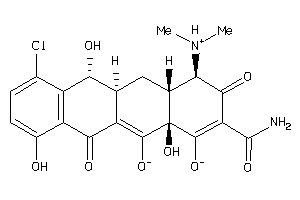
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-2-O |
Mycobacterium Smegmatis (cluster #2 Of 4), Other |
Other |
5920 |
0.23 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
794 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.38 |
-2.26 |
-63.05 |
6 |
10 |
-1 |
184 |
463.85 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
-0.93 |
-2.27 |
-62.34 |
6 |
10 |
-1 |
184 |
463.85 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.38 |
-1.28 |
-142.66 |
5 |
10 |
-2 |
187 |
462.842 |
1 |
↓
|
|
|
|
Analogs
-
19868839
-
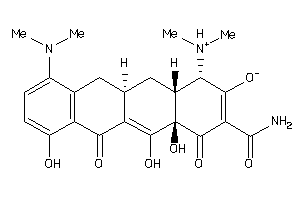
-
6566166
-
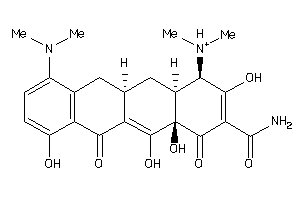
-
8691805
-
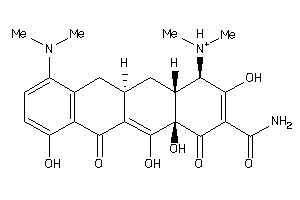
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.23 |
3.53 |
-52.8 |
6 |
10 |
0 |
169 |
457.483 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.23 |
2.49 |
-137.36 |
4 |
10 |
-2 |
170 |
455.467 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.96 |
-0.02 |
-17.13 |
6 |
10 |
0 |
165 |
457.483 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
538273
-
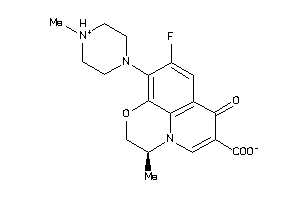
-
22056311
-
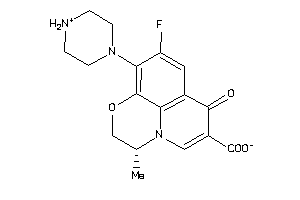
-
22056381
-
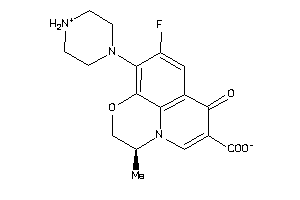
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.26 |
10.08 |
-92.39 |
1 |
7 |
0 |
79 |
361.373 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.26 |
7.74 |
-63.35 |
0 |
7 |
-1 |
78 |
360.365 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.01 |
8.22 |
-73.63 |
2 |
7 |
1 |
82 |
362.381 |
1 |
↓
|
|
|
|
Analogs
-
22056311
-

-
22056381
-

-
537891
-
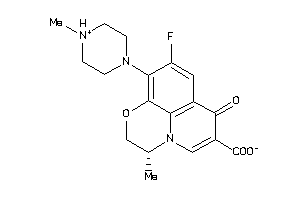
Draw
Identity
99%
90%
80%
70%
Vendors
And 69 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.26 |
9.75 |
-92.26 |
1 |
7 |
0 |
79 |
361.373 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.01 |
7.88 |
-73.44 |
2 |
7 |
1 |
82 |
362.381 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.26 |
7.41 |
-63.01 |
0 |
7 |
-1 |
78 |
360.365 |
2 |
↓
|
|
|
|
|