|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 66 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
10000 |
0.64 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.43 |
3.02 |
-10.14 |
3 |
5 |
0 |
80 |
169.575 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.43 |
2.48 |
-37.03 |
2 |
5 |
-1 |
79 |
168.567 |
0 |
↓
|
|
|
|
Analogs
-
582580
-
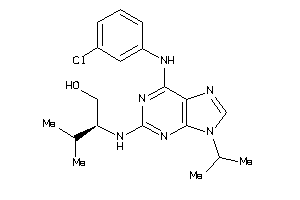
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.38 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.38 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.38 |
Binding ≤ 10μM
|
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.39 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.39 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.38 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Binding ≤ 10μM
|
|
CDK4-1-E |
Cyclin-dependent Kinase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
850 |
0.31 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
75 |
0.37 |
Binding ≤ 10μM
|
|
INSR-1-E |
Insulin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
MK03-1-E |
MAP Kinase ERK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
PAK4-1-E |
Serine/threonine-protein Kinase PAK 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
130 |
0.36 |
Binding ≤ 10μM
|
|
Q17066-1-E |
Cdc2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.33 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.34 |
Binding ≤ 10μM
|
|
CDK1-1-F |
Cyclin-dependent Kinase 1 (cluster #1 Of 1), Fungal |
Fungi |
80 |
0.37 |
Binding ≤ 10μM
|
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
850 |
0.31 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
9200 |
0.26 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
9000 |
0.26 |
Functional ≤ 10μM
|
|
Z80874-7-O |
CEM (T-cell Leukemia) (cluster #7 Of 7), Other |
Other |
7400 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.28 |
10.24 |
-10.66 |
3 |
7 |
0 |
88 |
388.903 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
7996842
-
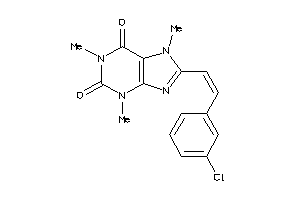
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
9.55 |
-11.56 |
0 |
6 |
0 |
62 |
330.775 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
|
CCNB-1-E |
G2/mitotic-specific Cyclin B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM |
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM |
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM |
|
CDK1-4-E |
Cyclin-dependent Kinase 1 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
|
CDK2-4-E |
Cyclin-dependent Kinase 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.35 |
Binding ≤ 10μM
|
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
7000 |
0.33 |
Binding ≤ 10μM
|
|
Z104296-2-O |
Cyclin-dependent Kinase 1/cyclin B1 (cluster #2 Of 2), Other |
Other |
7000 |
0.33 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
8000 |
0.32 |
Functional ≤ 10μM
|
|
Z80156-9-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #9 Of 12), Other |
Other |
50 |
0.46 |
Functional ≤ 10μM
|
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
57 |
0.46 |
Functional ≤ 10μM
|
|
Z80783-1-O |
COLO1 (cluster #1 Of 1), Other |
Other |
87 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
6.57 |
-12.73 |
3 |
7 |
0 |
88 |
298.35 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.04 |
6.85 |
-30.42 |
4 |
7 |
1 |
89 |
299.358 |
6 |
↓
|
|
|
|
Analogs
-
4475112
-
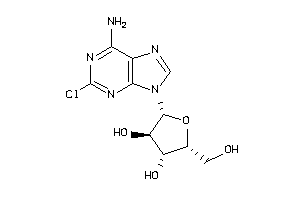
-
4536052
-
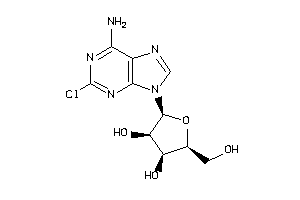
-
4536053
-
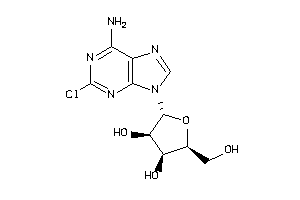
-
4536054
-
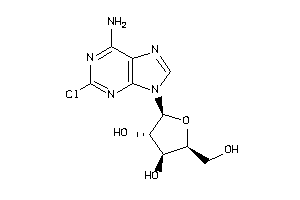
-
4536057
-
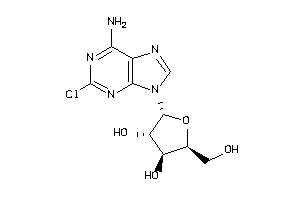
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
18 |
0.54 |
Binding ≤ 10μM |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
20 |
0.54 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
76 |
0.50 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
920 |
0.42 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.49 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3200 |
0.38 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Functional ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.44 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 5), Other |
Other |
920 |
0.42 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
7943 |
0.36 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
620 |
0.43 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
10000 |
0.35 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
3000 |
0.39 |
Functional ≤ 10μM
|
|
Z80945-1-O |
HEp-2 (Laryngeal Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
3000 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.14 |
-4.58 |
-14.68 |
5 |
9 |
0 |
140 |
301.69 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
6600 |
0.66 |
Functional ≤ 10μM
|
|
Z80945-3-O |
HEp-2 (Laryngeal Carcinoma Cells) (cluster #3 Of 4), Other |
Other |
2000 |
0.73 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.61 |
-3.1 |
-41.91 |
3 |
7 |
-1 |
116 |
151.109 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.61 |
-3.11 |
-108.43 |
2 |
7 |
-2 |
115 |
150.101 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.61 |
-3.21 |
-42.03 |
3 |
7 |
-1 |
116 |
151.109 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
7480 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.29 |
0.96 |
-12.58 |
3 |
6 |
0 |
87 |
151.129 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
4400 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
11.2 |
-41.22 |
0 |
5 |
-1 |
57 |
274.307 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
3.56 |
11.83 |
-14.48 |
1 |
5 |
0 |
55 |
275.315 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q54A96-1-E |
Dihydroorotate Dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
47 |
0.49 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
79 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
11.72 |
-14.33 |
1 |
5 |
0 |
55 |
275.315 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1185 |
0.52 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
6310 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
7.78 |
-9.41 |
1 |
5 |
0 |
56 |
217.276 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
4200 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
9.41 |
-35.03 |
0 |
5 |
-1 |
57 |
303.143 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
3.21 |
10.09 |
-13.55 |
1 |
5 |
0 |
55 |
304.151 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
6400 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
9.47 |
-39.56 |
0 |
5 |
-1 |
57 |
238.274 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
2.85 |
10.15 |
-13.59 |
1 |
5 |
0 |
55 |
239.282 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
3800 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
1.88 |
-11.01 |
1 |
6 |
0 |
64 |
309.251 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
8500 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
9.98 |
-12.54 |
1 |
5 |
0 |
55 |
259.7 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTR1-1-E |
Pteridine Reductase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
8822 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
2.67 |
-11.38 |
6 |
7 |
0 |
130 |
253.269 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.82 |
3.17 |
-25.15 |
7 |
7 |
1 |
131 |
254.277 |
1 |
↓
|
|
|
|
Analogs
-
1319816
-
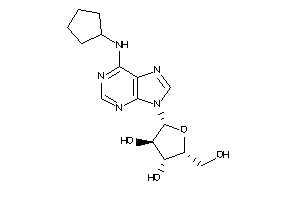
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
22 |
0.45 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
857 |
0.35 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
580 |
0.36 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
72 |
0.42 |
Binding ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.45 |
Binding ≤ 10μM
|
|
ADCY3-1-E |
Adenylate Cyclase Type III (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.45 |
Binding ≤ 10μM
|
|
ADCY4-1-E |
Adenylate Cyclase Type IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.45 |
Binding ≤ 10μM
|
|
ADCY5-1-E |
Adenylate Cyclase Type V (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.45 |
Binding ≤ 10μM
|
|
ADCY6-1-E |
Adenylate Cyclase Type VI (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.45 |
Binding ≤ 10μM
|
|
ADCY8-2-E |
Adenylate Cyclase Type VIII (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.45 |
Binding ≤ 10μM
|
|
Q4VQ11-1-E |
A1 Adenosine Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
810 |
0.36 |
Binding ≤ 10μM
|
|
S29A1-1-E |
Equilibrative Nucleoside Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.35 |
Binding ≤ 10μM
|
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Functional ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Functional ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.43 |
Functional ≤ 10μM
|
|
AA3R-1-E |
Adenosine A3 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7980 |
0.30 |
Functional ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Functional ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
10000 |
0.29 |
Functional ≤ 10μM
|
|
Z50512-5-O |
Cavia Porcellus (cluster #5 Of 7), Other |
Other |
420 |
0.37 |
Functional ≤ 10μM |
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
12 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
-0.24 |
-14.3 |
4 |
9 |
0 |
126 |
335.364 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.58 |
3.49 |
-22.11 |
4 |
5 |
0 |
83 |
167.197 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.14 |
3.08 |
-52.15 |
3 |
5 |
-1 |
80 |
166.189 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
39334986
-
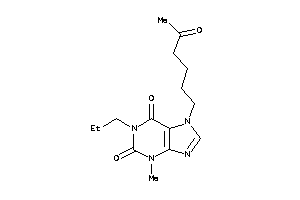
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
9.8 |
-14.83 |
0 |
7 |
0 |
79 |
306.366 |
7 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 68 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100081-3-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #3 Of 4), Other |
Other |
150 |
0.96 |
Functional ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
2000 |
0.80 |
Functional ≤ 10μM
|
|
Z80064-3-O |
CCRF-CEM (T-cell Leukemia) (cluster #3 Of 9), Other |
Other |
1000 |
0.84 |
Functional ≤ 10μM
|
|
Z80068-2-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #2 Of 5), Other |
Other |
1000 |
0.84 |
Functional ≤ 10μM
|
|
Z80125-5-O |
DU-145 (Prostate Carcinoma) (cluster #5 Of 9), Other |
Other |
2000 |
0.80 |
Functional ≤ 10μM
|
|
Z80193-6-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #6 Of 12), Other |
Other |
24 |
1.07 |
Functional ≤ 10μM
|
|
Z80224-4-O |
MCF7 (Breast Carcinoma Cells) (cluster #4 Of 14), Other |
Other |
5800 |
0.73 |
Functional ≤ 10μM
|
|
Z80295-4-O |
MT4 (Lymphocytes) (cluster #4 Of 8), Other |
Other |
100 |
0.98 |
Functional ≤ 10μM
|
|
Z80936-3-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #3 Of 4), Other |
Other |
7 |
1.14 |
Functional ≤ 10μM
|
|
Z81020-5-O |
HepG2 (Hepatoblastoma Cells) (cluster #5 Of 8), Other |
Other |
8000 |
0.71 |
Functional ≤ 10μM
|
|
Z81247-5-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #5 Of 9), Other |
Other |
4100 |
0.75 |
Functional ≤ 10μM
|
|
Z80178-1-O |
J774.A1 (Macrophage Cells) (cluster #1 Of 2), Other |
Other |
3 |
1.19 |
ADME/T ≤ 10μM
|
|
Z80936-3-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #3 Of 4), Other |
Other |
7 |
1.14 |
ADME/T ≤ 10μM
|
|
Z81244-2-O |
J774 (Macrophage Cells) (cluster #2 Of 3), Other |
Other |
3 |
1.19 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
4.06 |
-18.68 |
2 |
4 |
0 |
57 |
152.182 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.34 |
3.67 |
-44.86 |
1 |
4 |
-1 |
54 |
151.174 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.34 |
3.66 |
-43.68 |
1 |
4 |
-1 |
54 |
151.174 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 63 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
4200 |
0.75 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.73 |
1.96 |
-19.54 |
2 |
5 |
0 |
74 |
136.114 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.27 |
-0.19 |
-46.35 |
1 |
5 |
-1 |
78 |
135.106 |
0 |
↓
|
|
|
|
Analogs
-
602192
-
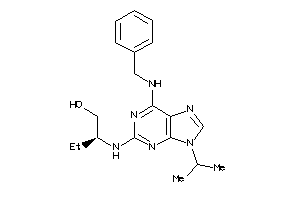
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.30 |
Binding ≤ 10μM
|
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNH-1-E |
Cyclin H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
540 |
0.34 |
Binding ≤ 10μM |
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
950 |
0.32 |
Binding ≤ 10μM |
|
CCNT2-1-E |
Cyclin-T2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
810 |
0.33 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CDK16-1-E |
Serine/threonine-protein Kinase PCTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
990 |
0.32 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.38 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.37 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.35 |
Binding ≤ 10μM
|
|
CDK7-1-E |
Cyclin-dependent Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.33 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
950 |
0.32 |
Binding ≤ 10μM |
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.31 |
Binding ≤ 10μM
|
|
CLK2-1-E |
Dual Specificity Protein Kinase CLK2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CLK4-1-E |
Dual Specificity Protein Kinase CLK4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.29 |
Binding ≤ 10μM
|
|
DYR1A-3-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.29 |
Binding ≤ 10μM
|
|
DYR1B-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.32 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
260 |
0.35 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.35 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.30 |
Binding ≤ 10μM
|
|
KC1G2-1-E |
Casein Kinase I Gamma 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.32 |
Binding ≤ 10μM
|
|
KC1G3-1-E |
Casein Kinase I Isoform Gamma-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.30 |
Binding ≤ 10μM
|
|
KS6A2-1-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.30 |
Binding ≤ 10μM
|
|
MAT1-1-E |
CDK-activating Kinase Assembly Factor MAT1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
540 |
0.34 |
Binding ≤ 10μM |
|
PAK4-1-E |
Serine/threonine-protein Kinase PAK 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.28 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.31 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
280 |
0.35 |
Binding ≤ 10μM |
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
940 |
0.32 |
Binding ≤ 10μM
|
|
Z104296-2-O |
Cyclin-dependent Kinase 1/cyclin B1 (cluster #2 Of 2), Other |
Other |
450 |
0.34 |
Binding ≤ 10μM
|
|
Z104299-1-O |
Cyclin-dependent Kinase 2/cyclin E1 (cluster #1 Of 1), Other |
Other |
220 |
0.36 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
6310 |
0.28 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
6000 |
0.28 |
Functional ≤ 10μM
|
|
Z80390-10-O |
PC-3 (Prostate Carcinoma Cells) (cluster #10 Of 10), Other |
Other |
8000 |
0.27 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
5000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
9.65 |
-10.26 |
3 |
7 |
0 |
88 |
354.458 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.65 |
9.9 |
-28.83 |
4 |
7 |
1 |
89 |
355.466 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.65 |
9.47 |
-25.05 |
4 |
7 |
1 |
89 |
355.466 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100081-3-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #3 Of 4), Other |
Other |
230 |
0.49 |
Functional ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.50 |
7.86 |
-28.67 |
1 |
9 |
0 |
118 |
277.269 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.50 |
7.39 |
-51.02 |
0 |
9 |
-1 |
116 |
276.261 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
7.88 |
-10.35 |
2 |
6 |
0 |
82 |
285.694 |
1 |
↓
|
|