|
|
Analogs
-
4096465
-
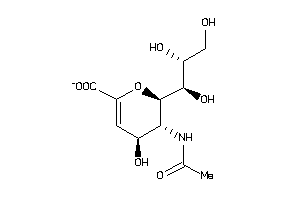
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.01 |
-9.78 |
-54.77 |
5 |
9 |
-1 |
159 |
290.248 |
5 |
↓
|
|
|
|
Analogs
-
4096465
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.01 |
-9.3 |
-62.44 |
5 |
9 |
-1 |
159 |
290.248 |
5 |
↓
|
|
|
|
Analogs
-
4096465
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.01 |
-10.47 |
-53.76 |
5 |
9 |
-1 |
159 |
290.248 |
5 |
↓
|
|
|
|
Analogs
-
4096465
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.01 |
-9.58 |
-59.1 |
5 |
9 |
-1 |
159 |
290.248 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A8K327-1-E |
Sialidase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM |
|
NEUR1-1-E |
Sialidase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM |
|
NEUR2-1-E |
Sialidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM |
|
NEUR3-1-E |
Sialidase-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.31 |
Binding ≤ 10μM
|
|
NEUR4-1-E |
Sialidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.34 |
Binding ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Binding ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 1), Other |
Other |
5 |
0.51 |
Binding ≤ 10μM
|
|
Z100811-1-O |
Influenza A Virus (H5N1) (cluster #1 Of 1), Other |
Other |
2800 |
0.34 |
Functional ≤ 10μM
|
|
Z101821-1-O |
H2N2 Subtype (cluster #1 Of 3), Other |
Other |
760 |
0.37 |
Functional ≤ 10μM
|
|
Z101822-1-O |
H3N2 Subtype (cluster #1 Of 2), Other |
Other |
680 |
0.38 |
Functional ≤ 10μM
|
|
Z50142-2-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #2 Of 2), Other |
Other |
11 |
0.48 |
Functional ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
90 |
0.43 |
Functional ≤ 10μM
|
|
Z80249-1-O |
MDCK (Kidney Cells) (cluster #1 Of 2), Other |
Other |
40 |
0.45 |
Functional ≤ 10μM |
|
A5Z252-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
1 |
0.55 |
Binding ≤ 10μM
|
|
B4URF0-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
4 |
0.51 |
Binding ≤ 10μM
|
|
C4LRQ6-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
1 |
0.55 |
Binding ≤ 10μM
|
|
NRAM-1-V |
Neuraminidase (cluster #1 Of 2), Viral |
Viruses |
3 |
0.52 |
Binding ≤ 10μM
|
|
Q2LFS1-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
1 |
0.55 |
Binding ≤ 10μM
|
|
Q6Q793-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
1 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.64 |
-6.67 |
-67.62 |
9 |
11 |
0 |
205 |
332.313 |
6 |
↓
|
|
|
|
Analogs
-
6777826
-
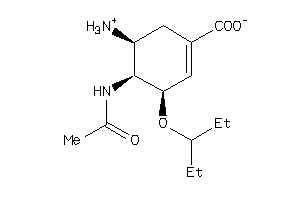
-
6777828
-
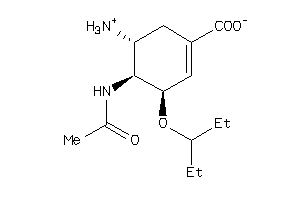
-
6777829
-
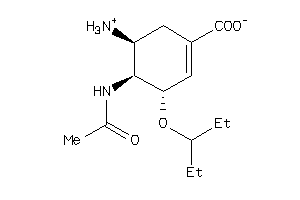
-
6777830
-
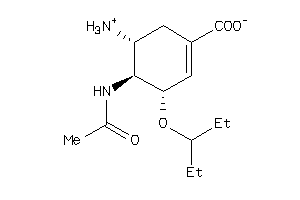
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A8K327-1-E |
Sialidase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
A8K327-1-E |
Sialidase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM
|
|
NEUR1-1-E |
Sialidase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
NEUR1-1-E |
Sialidase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM
|
|
NEUR2-1-E |
Sialidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
NEUR2-1-E |
Sialidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM
|
|
NEUR4-1-E |
Sialidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
4 |
0.59 |
Binding ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 1), Other |
Other |
2 |
0.61 |
Binding ≤ 10μM
|
|
Z101821-1-O |
H2N2 Subtype (cluster #1 Of 3), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z101822-1-O |
H3N2 Subtype (cluster #1 Of 2), Other |
Other |
1 |
0.63 |
Functional ≤ 10μM
|
|
Z50142-2-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #2 Of 2), Other |
Other |
8300 |
0.36 |
Functional ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
15 |
0.55 |
Functional ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
6250 |
0.36 |
Functional ≤ 10μM
|
|
Z80249-2-O |
MDCK (Kidney Cells) (cluster #2 Of 2), Other |
Other |
2240 |
0.40 |
ADME/T ≤ 10μM |
|
NRAM-1-V |
Neuraminidase (cluster #1 Of 2), Viral |
Viruses |
5 |
0.58 |
Binding ≤ 10μM
|
|
NRAM-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
4 |
0.59 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.14 |
-2.58 |
-79.64 |
4 |
6 |
0 |
106 |
284.356 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
11592802
-
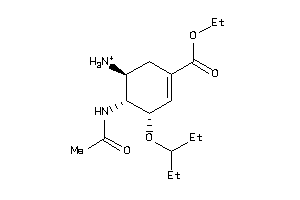
-
36451498
-
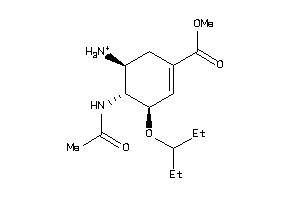
-
3874571
-
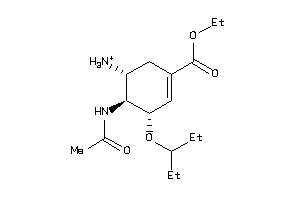
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101821-1-O |
H2N2 Subtype (cluster #1 Of 3), Other |
Other |
1450 |
0.37 |
Functional ≤ 10μM
|
|
Z101822-1-O |
H3N2 Subtype (cluster #1 Of 2), Other |
Other |
260 |
0.42 |
Functional ≤ 10μM
|
|
Z50142-2-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #2 Of 2), Other |
Other |
8600 |
0.32 |
Functional ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
2600 |
0.36 |
Functional ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
A5Z252-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
8 |
0.52 |
Binding ≤ 10μM
|
|
C4LRQ6-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
1 |
0.57 |
Binding ≤ 10μM
|
|
NRAM-1-V |
Neuraminidase (cluster #1 Of 2), Viral |
Viruses |
2 |
0.55 |
Binding ≤ 10μM
|
|
Q2LFS1-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
5 |
0.53 |
Binding ≤ 10μM
|
|
Q6Q793-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.85 |
5.84 |
-62.6 |
4 |
6 |
1 |
92 |
313.418 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.85 |
5.55 |
-12.89 |
3 |
6 |
0 |
91 |
312.41 |
8 |
↓
|
|
|
|
Analogs
-
34022665
-
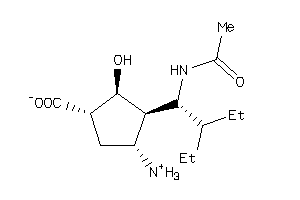
-
34031444
-
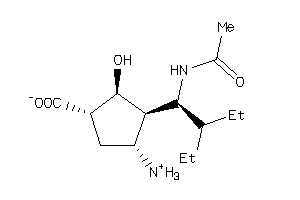
-
40709762
-
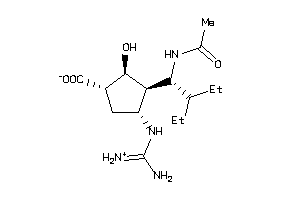
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.10 |
2.44 |
-59.87 |
7 |
8 |
0 |
153 |
328.413 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.77 |
23.55 |
-33.73 |
0 |
5 |
1 |
56 |
588.683 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABI-1-B |
Enoyl-[acyl-carrier-protein] Reductase (cluster #1 Of 2), Bacterial |
Bacteria |
7050 |
0.34 |
Binding ≤ 10μM
|
|
NANH-1-B |
Sialidase (cluster #1 Of 1), Bacterial |
Bacteria |
4300 |
0.36 |
Binding ≤ 10μM
|
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1660 |
0.39 |
Binding ≤ 10μM
|
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8900 |
0.34 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
9570 |
0.33 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CD38-1-E |
Lymphocyte Differentiation Antigen CD38 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8200 |
0.34 |
Binding ≤ 10μM |
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.41 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
800 |
0.41 |
Binding ≤ 10μM
|
|
LGUL-2-E |
Glyoxalase I (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7700 |
0.34 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.37 |
Binding ≤ 10μM
|
|
NOX4-1-E |
NADPH Oxidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
850 |
0.40 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.38 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.41 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.35 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.41 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1450 |
0.39 |
Functional ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
890 |
0.40 |
ADME/T ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3370 |
0.36 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
56 |
0.48 |
ADME/T ≤ 10μM
|
|
Z104294-2-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #2 Of 2), Other |
Other |
3800 |
0.36 |
Binding ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
9600 |
0.33 |
Functional ≤ 10μM
|
|
Z50607-4-O |
Human Immunodeficiency Virus 1 (cluster #4 Of 10), Other |
Other |
10000 |
0.33 |
Functional ≤ 10μM |
|
Z50652-3-O |
Influenza A Virus (cluster #3 Of 4), Other |
Other |
6820 |
0.34 |
Functional ≤ 10μM
|
|
Z80150-2-O |
H9c2 (Cardiomyoblast Cells) (cluster #2 Of 2), Other |
Other |
5530 |
0.35 |
Functional ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
7500 |
0.34 |
Functional ≤ 10μM
|
|
Z80420-1-O |
RBL-2H3 (Basophilic Leukemia Cells) (cluster #1 Of 2), Other |
Other |
5800 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
-0.94 |
-14.61 |
4 |
6 |
0 |
111 |
286.239 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
4245717
-
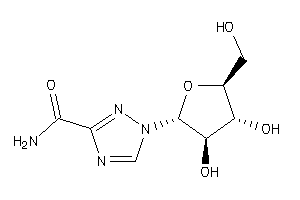
-
6603353
-
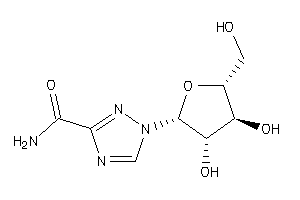
Draw
Identity
99%
90%
80%
70%
Vendors
-
-
-
- API0004062
- API0009704
- CCH0012019
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100497-1-O |
Bovine Viral Diarrhea Virus (cluster #1 Of 3), Other |
Other |
7000 |
0.42 |
Functional ≤ 10μM
|
|
Z100811-1-O |
Influenza A Virus (H5N1) (cluster #1 Of 1), Other |
Other |
9400 |
0.41 |
Functional ≤ 10μM
|
|
Z101822-1-O |
H3N2 Subtype (cluster #1 Of 2), Other |
Other |
9000 |
0.42 |
Functional ≤ 10μM
|
|
Z50142-1-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #1 Of 2), Other |
Other |
9700 |
0.41 |
Functional ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
9000 |
0.42 |
Functional ≤ 10μM
|
|
Z50525-1-O |
Tacaribe Virus (cluster #1 Of 1), Other |
Other |
10000 |
0.41 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
8300 |
0.42 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
10000 |
0.41 |
Functional ≤ 10μM
|
|
Z50641-1-O |
West Nile Virus (cluster #1 Of 2), Other |
Other |
1100 |
0.49 |
Functional ≤ 10μM
|
|
Z50651-1-O |
Vesicular Stomatitis Virus (cluster #1 Of 2), Other |
Other |
10000 |
0.41 |
Functional ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
9800 |
0.41 |
Functional ≤ 10μM |
|
Z50678-2-O |
Respiratory Syncytial Virus (cluster #2 Of 2), Other |
Other |
7000 |
0.42 |
Functional ≤ 10μM
|
|
Z80040-1-O |
BHK-21 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
1100 |
0.49 |
Functional ≤ 10μM
|
|
Z80249-1-O |
MDCK (Kidney Cells) (cluster #1 Of 2), Other |
Other |
4900 |
0.44 |
Functional ≤ 10μM
|
|
Z80285-1-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
5700 |
0.43 |
Functional ≤ 10μM |
|
Z80823-1-O |
E6SM (Embryonic Skin-muscle Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
5 |
0.68 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1500 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.77 |
-8.13 |
-17.41 |
5 |
9 |
0 |
144 |
244.207 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.52 |
-3.28 |
-40.6 |
2 |
5 |
-1 |
92 |
156.096 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.98 |
-2.18 |
-20.54 |
3 |
5 |
0 |
89 |
157.104 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 48 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
|
AA2AR-3-E |
Adenosine A2a Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7580 |
0.36 |
Binding ≤ 10μM
|
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5900 |
0.37 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6670 |
0.36 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
1700 |
0.40 |
Binding ≤ 10μM
|
|
BGLR-1-E |
Beta-glucuronidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
CDK6-1-E |
Cyclin-dependent Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.40 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.53 |
Binding ≤ 10μM
|
|
CSK21-2-E |
Casein Kinase II Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Binding ≤ 10μM
|
|
CSK2B-3-E |
Casein Kinase II Beta (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4290 |
0.38 |
Binding ≤ 10μM
|
|
DHB1-1-E |
Estradiol 17-beta-dehydrogenase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.43 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
660 |
0.43 |
Binding ≤ 10μM
|
|
ESR2-4-E |
Estrogen Receptor Beta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
660 |
0.43 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GBRA6-6-E |
GABA Receptor Alpha-6 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
MRP1-1-E |
Multidrug Resistance-associated Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.39 |
Binding ≤ 10μM
|
|
NOX4-1-E |
NADPH Oxidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1130 |
0.42 |
Binding ≤ 10μM
|
|
P90584-1-E |
Protein Kinase Pfmrk (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.36 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.43 |
Binding ≤ 10μM
|
|
ANDR-1-E |
Androgen Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5200 |
0.37 |
Functional ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
427 |
0.45 |
ADME/T ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
795 |
0.43 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
64 |
0.50 |
ADME/T ≤ 10μM
|
|
Z104294-2-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #2 Of 2), Other |
Other |
1600 |
0.41 |
Binding ≤ 10μM
|
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
770 |
0.43 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3400 |
0.38 |
Functional ≤ 10μM
|
|
Z50607-3-O |
Human Immunodeficiency Virus 1 (cluster #3 Of 10), Other |
Other |
9000 |
0.35 |
Functional ≤ 10μM |
|
Z50652-3-O |
Influenza A Virus (cluster #3 Of 4), Other |
Other |
4740 |
0.37 |
Functional ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
7700 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
1.12 |
-12.82 |
3 |
5 |
0 |
91 |
270.24 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.72 |
1.4 |
-45.92 |
2 |
5 |
-1 |
94 |
269.232 |
1 |
↓
|
|
|
|
Analogs
-
12659498
-
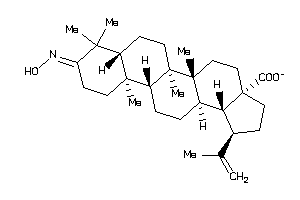
-
8762630
-
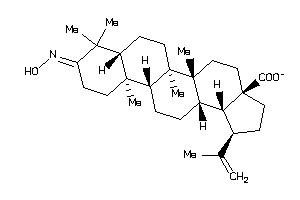
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
2200 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.31 |
11.84 |
-51.44 |
1 |
4 |
-1 |
73 |
468.702 |
2 |
↓
|
|
|
|
Analogs
-
8762630
-

-
8762634
-
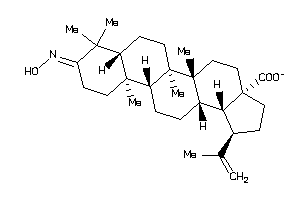
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.31 |
11.6 |
-54 |
1 |
4 |
-1 |
73 |
468.702 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,5aR,5bR,7aR,9E,11aR,11bR,13aS,13bS)-9-hydroxyimino-1-isopropenyl-5a,5b,8,8,11a-pentamethyl-2
(1R,3aS,5aR,5bR,7aR,9E,11aR,11bR…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
2200 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.31 |
12.22 |
-53.37 |
1 |
4 |
-1 |
73 |
468.702 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.31 |
10.16 |
-6.57 |
2 |
4 |
0 |
70 |
469.71 |
2 |
↓
|
|
|
|
Analogs
-
8762249
-
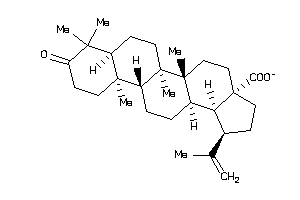
-
8762252
-
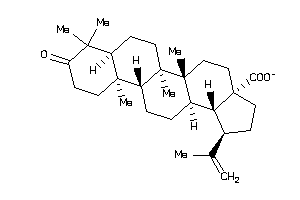
-
8835967
-
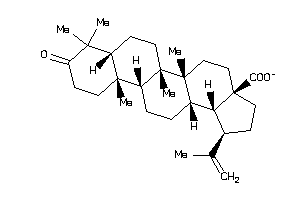
-
8835968
-
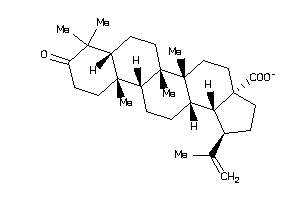
-
8952537
-
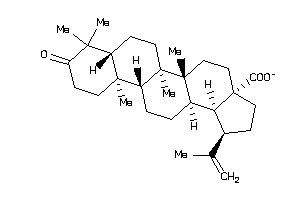
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4710 |
0.23 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z100499-2-O |
SARS Coronavirus (cluster #2 Of 2), Other |
Other |
630 |
0.26 |
Functional ≤ 10μM
|
|
Z103205-2-O |
A431 (cluster #2 Of 4), Other |
Other |
3260 |
0.23 |
Functional ≤ 10μM
|
|
Z50602-3-O |
Human Herpesvirus 1 (cluster #3 Of 5), Other |
Other |
2500 |
0.24 |
Functional ≤ 10μM
|
|
Z50652-2-O |
Influenza A Virus (cluster #2 Of 4), Other |
Other |
5700 |
0.22 |
Functional ≤ 10μM
|
|
Z80186-4-O |
K562 (Erythroleukemia Cells) (cluster #4 Of 11), Other |
Other |
6000 |
0.22 |
Functional ≤ 10μM
|
|
Z80928-4-O |
HCT-116 (Colon Carcinoma Cells) (cluster #4 Of 9), Other |
Other |
2610 |
0.24 |
Functional ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
2980 |
0.23 |
Functional ≤ 10μM
|
|
Z81252-4-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #4 Of 11), Other |
Other |
9770 |
0.21 |
Functional ≤ 10μM
|
|
Z80186-2-O |
K562 (Erythroleukemia Cells) (cluster #2 Of 3), Other |
Other |
6000 |
0.22 |
ADME/T ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 3), Other |
Other |
3800 |
0.23 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.86 |
14.43 |
-48.14 |
0 |
3 |
-1 |
57 |
453.687 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.86 |
12.66 |
-7.5 |
1 |
3 |
0 |
54 |
454.695 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
3831612
-
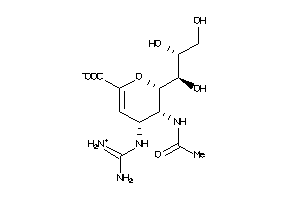
-
3831613
-
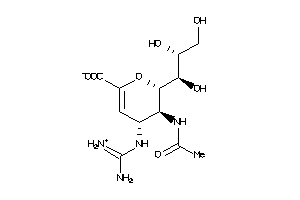
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.11 |
-10.28 |
-72.57 |
8 |
11 |
0 |
191 |
346.34 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.43 |
-10.24 |
-53.08 |
7 |
12 |
-1 |
209 |
365.315 |
7 |
↓
|
|
|
|
Analogs
-
3206938
-
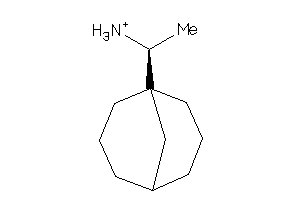
-
3206939
-
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
-1.33 |
-39.51 |
3 |
1 |
1 |
27 |
180.315 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A8K327-1-E |
Sialidase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
A8K327-1-E |
Sialidase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
NEUR1-1-E |
Sialidase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
NEUR1-1-E |
Sialidase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
NEUR2-1-E |
Sialidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
NEUR2-1-E |
Sialidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
NEUR4-1-E |
Sialidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
NEUR4-1-E |
Sialidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
NRAM-1-V |
Neuraminidase (cluster #1 Of 2), Viral |
Viruses |
410 |
0.45 |
Binding ≤ 10μM |
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
1500 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.50 |
-11.28 |
-76.12 |
7 |
9 |
0 |
166 |
290.272 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-3-O |
Trypanosoma Brucei (cluster #3 Of 6), Other |
Other |
5280 |
0.49 |
Functional ≤ 10μM |
|
Z50652-2-O |
Influenza A Virus (cluster #2 Of 4), Other |
Other |
160 |
0.63 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
6.43 |
-42.28 |
2 |
1 |
1 |
17 |
206.353 |
0 |
↓
|
|
|
|
Analogs
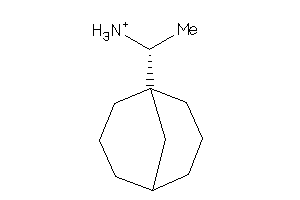
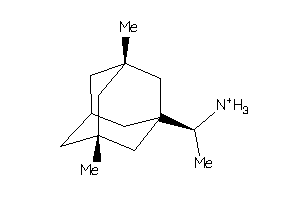
-
4159387
-
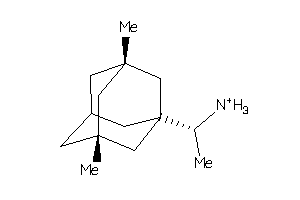
-
4159388
-
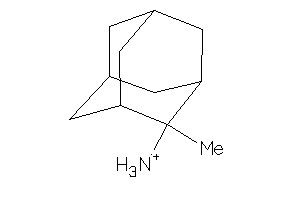
-
40865734
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 87 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
5.01 |
-42.88 |
3 |
1 |
1 |
28 |
180.315 |
1 |
↓
|
|
|
|
|