|
|
Analogs
-
4212843
-
-
607790
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q862F3-2-E |
Tubulin Alpha Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM |
|
Q862L2-2-E |
Tubulin Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM |
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.27 |
Binding ≤ 10μM
|
|
TBB-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1260 |
0.28 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.29 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.29 |
Binding ≤ 10μM
|
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.30 |
Binding ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.30 |
Binding ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.30 |
Binding ≤ 10μM
|
|
TBG1-1-E |
Tubulin Gamma-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5750 |
0.25 |
Binding ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.24 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.24 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.29 |
Functional ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.28 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.29 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.24 |
Functional ≤ 10μM
|
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.26 |
Functional ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.26 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
22 |
0.37 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM |
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
53 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
1400 |
0.28 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
18 |
0.37 |
Functional ≤ 10μM |
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
12 |
0.38 |
Functional ≤ 10μM
|
|
Z80024-1-O |
A9 (Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.36 |
Functional ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
31 |
0.36 |
Functional ≤ 10μM
|
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
220 |
0.32 |
Functional ≤ 10μM |
|
Z80096-1-O |
CHO-TAX 5-6 (cluster #1 Of 2), Other |
Other |
140 |
0.33 |
Functional ≤ 10μM |
|
Z80097-1-O |
CHO-VV 3-2 (cluster #1 Of 2), Other |
Other |
620 |
0.30 |
Functional ≤ 10μM |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
190 |
0.32 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
10 |
0.39 |
Functional ≤ 10μM |
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
53 |
0.35 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
1 |
0.43 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
22 |
0.37 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1900 |
0.28 |
Functional ≤ 10μM |
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
15 |
0.38 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
800 |
0.29 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
10 |
0.39 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
33 |
0.36 |
Functional ≤ 10μM |
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM
|
|
Z80427-1-O |
RKO (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM |
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
71 |
0.35 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
30 |
0.36 |
Functional ≤ 10μM |
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
390 |
0.31 |
Functional ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
45 |
0.35 |
Functional ≤ 10μM
|
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
1113 |
0.29 |
Functional ≤ 10μM |
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
9 |
0.39 |
Functional ≤ 10μM
|
|
Z80720-1-O |
Burkitts Lymphoma Cells (cluster #1 Of 1), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
52 |
0.35 |
Functional ≤ 10μM |
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
3 |
0.41 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
8 |
0.39 |
Functional ≤ 10μM |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
40 |
0.36 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM |
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
70 |
0.35 |
Functional ≤ 10μM |
|
Z81054-2-O |
SF-268 (Glioblastoma Cells) (cluster #2 Of 3), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
17 |
0.38 |
Functional ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM |
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
93 |
0.34 |
Functional ≤ 10μM |
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
39 |
0.36 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
70 |
0.35 |
Functional ≤ 10μM
|
|
Z81281-1-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 5), Other |
Other |
13 |
0.38 |
Functional ≤ 10μM |
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM |
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
14 |
0.38 |
Functional ≤ 10μM |
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
140 |
0.33 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 3), Other |
Other |
530 |
0.30 |
ADME/T ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
522 |
0.30 |
ADME/T ≤ 10μM
|
|
Z80897-1-O |
H9 (T-lymphoid Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.39 |
ADME/T ≤ 10μM |
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.41 |
ADME/T ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 3), Other |
Other |
210 |
0.32 |
ADME/T ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
2 |
0.42 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
7.99 |
-20.74 |
1 |
7 |
0 |
83 |
399.443 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 75 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-3-E |
Thymidylate Synthase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.80 |
Binding ≤ 10μM
|
|
Z103205-4-O |
A431 (cluster #4 Of 4), Other |
Other |
18 |
1.20 |
Functional ≤ 10μM
|
|
Z103419-1-O |
UACC-812 (cluster #1 Of 2), Other |
Other |
3100 |
0.86 |
Functional ≤ 10μM
|
|
Z50425-14-O |
Plasmodium Falciparum (cluster #14 Of 22), Other |
Other |
7943 |
0.79 |
Functional ≤ 10μM
|
|
Z80035-6-O |
B16 (Melanoma Cells) (cluster #6 Of 7), Other |
Other |
8730 |
0.79 |
Functional ≤ 10μM
|
|
Z80054-2-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #2 Of 4), Other |
Other |
1500 |
0.91 |
Functional ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
5000 |
0.82 |
Functional ≤ 10μM
|
|
Z80088-4-O |
CHO (Ovarian Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.82 |
Functional ≤ 10μM
|
|
Z80101-1-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
1100 |
0.93 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
6000 |
0.81 |
Functional ≤ 10μM |
|
Z80125-6-O |
DU-145 (Prostate Carcinoma) (cluster #6 Of 9), Other |
Other |
4800 |
0.83 |
Functional ≤ 10μM
|
|
Z80136-3-O |
FM3A (Breast Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
180 |
1.05 |
Functional ≤ 10μM
|
|
Z80156-6-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #6 Of 12), Other |
Other |
9500 |
0.78 |
Functional ≤ 10μM
|
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
9450 |
0.78 |
Functional ≤ 10μM
|
|
Z80166-8-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #8 Of 12), Other |
Other |
7300 |
0.80 |
Functional ≤ 10μM
|
|
Z80186-7-O |
K562 (Erythroleukemia Cells) (cluster #7 Of 11), Other |
Other |
2400 |
0.87 |
Functional ≤ 10μM
|
|
Z80192-2-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #2 Of 4), Other |
Other |
8320 |
0.79 |
Functional ≤ 10μM
|
|
Z80193-8-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #8 Of 12), Other |
Other |
970 |
0.94 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
9600 |
0.78 |
Functional ≤ 10μM
|
|
Z80268-1-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2500 |
0.87 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3000 |
0.86 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
4400 |
0.83 |
Functional ≤ 10μM |
|
Z80362-5-O |
P388 (Lymphoma Cells) (cluster #5 Of 8), Other |
Other |
609 |
0.97 |
Functional ≤ 10μM |
|
Z80390-6-O |
PC-3 (Prostate Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
16 |
1.21 |
Functional ≤ 10μM
|
|
Z80485-4-O |
SK-MEL-28 (Melanoma Cells) (cluster #4 Of 6), Other |
Other |
4500 |
0.83 |
Functional ≤ 10μM
|
|
Z80490-4-O |
SK-MES-1 (cluster #4 Of 4), Other |
Other |
310 |
1.01 |
Functional ≤ 10μM
|
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
5700 |
0.82 |
Functional ≤ 10μM |
|
Z80526-3-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #3 Of 6), Other |
Other |
8100 |
0.79 |
Functional ≤ 10μM
|
|
Z80532-5-O |
T-24 (Bladder Carcinoma Cells) (cluster #5 Of 6), Other |
Other |
4100 |
0.84 |
Functional ≤ 10μM
|
|
Z80583-6-O |
Vero (Kidney Cells) (cluster #6 Of 7), Other |
Other |
6810 |
0.80 |
Functional ≤ 10μM
|
|
Z80641-1-O |
791T Cell Line (cluster #1 Of 1), Other |
Other |
5500 |
0.82 |
Functional ≤ 10μM
|
|
Z80653-2-O |
9L (Glioma Cells) (cluster #2 Of 2), Other |
Other |
1000 |
0.93 |
Functional ≤ 10μM
|
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
9200 |
0.78 |
Functional ≤ 10μM
|
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
3900 |
0.84 |
Functional ≤ 10μM |
|
Z80712-5-O |
T47D (Breast Carcinoma Cells) (cluster #5 Of 7), Other |
Other |
600 |
0.97 |
Functional ≤ 10μM
|
|
Z80724-1-O |
C170 Cell Line (cluster #1 Of 1), Other |
Other |
2000 |
0.89 |
Functional ≤ 10μM
|
|
Z80742-2-O |
C6 (Glioma Cells) (cluster #2 Of 3), Other |
Other |
6100 |
0.81 |
Functional ≤ 10μM
|
|
Z80874-5-O |
CEM (T-cell Leukemia) (cluster #5 Of 7), Other |
Other |
9230 |
0.78 |
Functional ≤ 10μM
|
|
Z80928-7-O |
HCT-116 (Colon Carcinoma Cells) (cluster #7 Of 9), Other |
Other |
8510 |
0.79 |
Functional ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 8), Other |
Other |
9200 |
0.78 |
Functional ≤ 10μM
|
|
Z81024-6-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #6 Of 8), Other |
Other |
8500 |
0.79 |
Functional ≤ 10μM |
|
Z81034-5-O |
A2780 (Ovarian Carcinoma Cells) (cluster #5 Of 10), Other |
Other |
7210 |
0.80 |
Functional ≤ 10μM
|
|
Z81041-1-O |
Prostatic Carcinoma Cells (cluster #1 Of 2), Other |
Other |
6400 |
0.81 |
Functional ≤ 10μM
|
|
Z81043-2-O |
N592 (cluster #2 Of 2), Other |
Other |
5400 |
0.82 |
Functional ≤ 10μM
|
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3600 |
0.85 |
Functional ≤ 10μM
|
|
Z81115-6-O |
KB (Squamous Cell Carcinoma) (cluster #6 Of 6), Other |
Other |
4460 |
0.83 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
6200 |
0.81 |
Functional ≤ 10μM |
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
4900 |
0.83 |
Functional ≤ 10μM
|
|
Z81247-7-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #7 Of 9), Other |
Other |
9700 |
0.78 |
Functional ≤ 10μM
|
|
Z81252-9-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #9 Of 11), Other |
Other |
9600 |
0.78 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
2700 |
0.87 |
Functional ≤ 10μM |
|
Z81331-3-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #3 Of 6), Other |
Other |
9000 |
0.78 |
Functional ≤ 10μM
|
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
9710 |
0.78 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 4), Other |
Other |
160 |
1.06 |
ADME/T ≤ 10μM
|
|
Z80192-2-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
7940 |
0.79 |
ADME/T ≤ 10μM
|
|
Z80193-4-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #4 Of 4), Other |
Other |
560 |
0.97 |
ADME/T ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
200 |
1.04 |
ADME/T ≤ 10μM
|
|
Z80901-2-O |
HaCaT (Keratinocytes) (cluster #2 Of 2), Other |
Other |
400 |
1.00 |
ADME/T ≤ 10μM
|
|
Z80928-2-O |
HCT-116 (Colon Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
2040 |
0.89 |
ADME/T ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 3), Other |
Other |
570 |
0.97 |
ADME/T ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
6610 |
0.81 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.59 |
-1.23 |
-13.33 |
2 |
4 |
0 |
66 |
130.078 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.13 |
-4 |
-48.58 |
1 |
4 |
-1 |
69 |
129.07 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.13 |
-3.14 |
-32.57 |
1 |
4 |
-1 |
69 |
129.07 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
308 |
0.35 |
ADME/T ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
92 |
0.38 |
ADME/T ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
30 |
0.41 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
9.68 |
-12.21 |
0 |
7 |
0 |
62 |
354.41 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-2-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
900 |
0.35 |
ADME/T ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
900 |
0.35 |
ADME/T ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 1), Other |
Other |
400 |
0.37 |
ADME/T ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
4000 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
5.63 |
-12.29 |
2 |
7 |
0 |
84 |
322.303 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
148 |
0.37 |
Binding ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
31 |
0.40 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
4 |
0.45 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
7.33 |
-99.64 |
4 |
6 |
2 |
76 |
352.438 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.38 |
6.03 |
-16.02 |
2 |
6 |
0 |
70 |
350.422 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.41 |
5.12 |
-38.36 |
3 |
6 |
1 |
75 |
351.43 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.35 |
Binding ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.35 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Binding ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.34 |
Binding ≤ 10μM |
|
Q25270-2-E |
Beta Tubulin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.35 |
Functional ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.38 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.38 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Functional ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4920 |
0.32 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.38 |
Functional ≤ 10μM
|
|
TBB3-3-E |
Tubulin Beta-3 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB4A-3-E |
Tubulin Beta-4 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB4B-3-E |
Tubulin Beta-2 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB8-3-E |
Tubulin Beta-8 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
Z100495-1-O |
BMEC (Brain Microvessel Endothelial Cells) (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z100742-1-O |
MeWo (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM |
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
188 |
0.41 |
Functional ≤ 10μM |
|
Z103208-1-O |
AU565 (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
|
Z103407-2-O |
SW1990 (cluster #2 Of 2), Other |
Other |
250 |
0.40 |
Functional ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
2000 |
0.35 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
18 |
0.47 |
Functional ≤ 10μM
|
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z80013-2-O |
A10 (cluster #2 Of 2), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80035-5-O |
B16 (Melanoma Cells) (cluster #5 Of 7), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z80064-4-O |
CCRF-CEM (T-cell Leukemia) (cluster #4 Of 9), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80071-2-O |
CEM-0 (T-cell Leukemia) (cluster #2 Of 2), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80099-3-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
2756 |
0.34 |
Functional ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
36 |
0.45 |
Functional ≤ 10μM
|
|
Z80121-1-O |
DMS-79 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
124 |
0.42 |
Functional ≤ 10μM
|
|
Z80125-9-O |
DU-145 (Prostate Carcinoma) (cluster #9 Of 9), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z80136-2-O |
FM3A (Breast Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
42 |
0.45 |
Functional ≤ 10μM |
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
94 |
0.43 |
Functional ≤ 10μM |
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM |
|
Z80193-1-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #1 Of 12), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80229-1-O |
MCF7-DOX (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80254-1-O |
MES-SA (Uterine Sarcoma Cells) (cluster #1 Of 2), Other |
Other |
13 |
0.48 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
82 |
0.43 |
Functional ≤ 10μM
|
|
Z80284-3-O |
MOLT-3 (T-lymphoblastic Leukemia Cells) (cluster #3 Of 3), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
16 |
0.47 |
Functional ≤ 10μM
|
|
Z80353-1-O |
OVCAR (cluster #1 Of 1), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
5400 |
0.32 |
Functional ≤ 10μM
|
|
Z80471-3-O |
SH-SY5 (Bone Marrow Neuroblastoma Cells) (cluster #3 Of 3), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM
|
|
Z80485-3-O |
SK-MEL-28 (Melanoma Cells) (cluster #3 Of 6), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM |
|
Z80555-1-O |
TSGH (cluster #1 Of 1), Other |
Other |
45 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80697-3-O |
Bel-7402 (Hepatoma Cells) (cluster #3 Of 3), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80792-1-O |
Colon 26 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z80798-1-O |
CPT30 Cell Line (cluster #1 Of 1), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z80847-3-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #3 Of 3), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80852-2-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z80874-2-O |
CEM (T-cell Leukemia) (cluster #2 Of 7), Other |
Other |
12 |
0.48 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80900-1-O |
HA22T Cell Line (cluster #1 Of 1), Other |
Other |
2708 |
0.34 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM
|
|
Z80967-2-O |
Hone-1 Cell Line (cluster #2 Of 2), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z81001-1-O |
TSGH 9201 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
38 |
0.45 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM
|
|
Z81024-5-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #5 Of 8), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z81065-3-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z81233-2-O |
M21 (Melanoma Cells) (cluster #2 Of 2), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
|
Z81252-7-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #7 Of 11), Other |
Other |
43 |
0.45 |
Functional ≤ 10μM |
|
Z81331-6-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #6 Of 6), Other |
Other |
23 |
0.46 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z100495-1-O |
BMEC (Brain Microvessel Endothelial Cells) (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
ADME/T ≤ 10μM
|
|
Z80013-2-O |
A10 (cluster #2 Of 2), Other |
Other |
23 |
0.46 |
ADME/T ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.52 |
ADME/T ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
ADME/T ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 4), Other |
Other |
2 |
0.53 |
ADME/T ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
ADME/T ≤ 10μM
|
|
Z81057-2-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #2 Of 5), Other |
Other |
5 |
0.51 |
ADME/T ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
3 |
0.52 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
5.32 |
-10.67 |
1 |
5 |
0 |
57 |
316.353 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
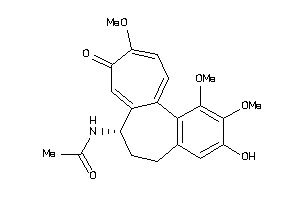
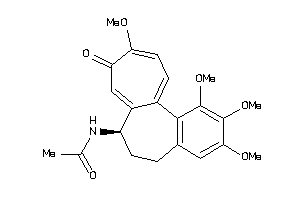
Popular Name:
[(2R,4aS,5R,10bS)-5-phenyl-9-(trifluoromethyl)-3,4,4a,5,6,10b-hexahydro-2H-pyrano[3,2-c]quinolin-2-y
[(2R,4aS,5R,10bS)-5-phenyl-9-(tr…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.41 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
4 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
7.3 |
-45.4 |
4 |
3 |
1 |
49 |
363.403 |
3 |
↓
|
|
|
|
Analogs
-
30780140
-
-
30780980
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
420 |
0.43 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
840 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
8.68 |
-4.14 |
1 |
2 |
0 |
21 |
279.383 |
1 |
↓
|
|
|
|
Analogs
-
30780184
-
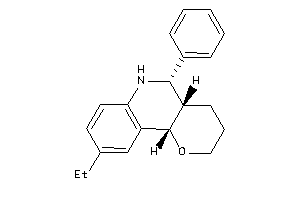
-
30780463
-
Draw
Identity
99%
90%
80%
70%
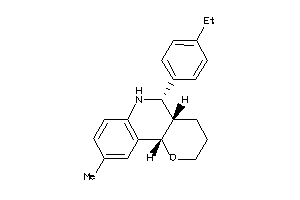
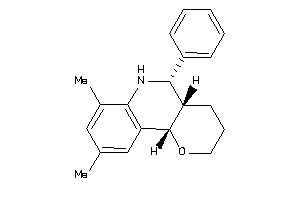
Popular Name:
(4aS,5R,10bS)-9-ethyl-5-phenyl-3,4,4a,5,6,10b-hexahydro-2H-pyrano[3,2-c]quinoline
(4aS,5R,10bS)-9-ethyl-5-phenyl-3…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.44 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
370 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.80 |
9.47 |
-3.99 |
1 |
2 |
0 |
21 |
293.41 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.41 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
1400 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
-1.35 |
-3.9 |
1 |
2 |
0 |
21 |
299.801 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80491-1-O |
SK-N-MC (Neuroepithelioma Cells) (cluster #1 Of 4), Other |
Other |
2880 |
0.37 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
1580 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
-0.14 |
-11.75 |
1 |
4 |
0 |
54 |
278.311 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.21 |
-5.18 |
0 |
2 |
0 |
26 |
225.506 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
355 |
0.27 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
355 |
0.27 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1904 |
0.24 |
Binding ≤ 10μM
|
|
PLK1-1-E |
Serine/threonine-protein Kinase PLK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.35 |
Binding ≤ 10μM
|
|
PLK2-1-E |
Serine/threonine-protein Kinase PLK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
238 |
0.28 |
Binding ≤ 10μM
|
|
PLK3-1-E |
Serine/threonine-protein Kinase PLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
450 |
0.27 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
587 |
0.26 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
219 |
0.28 |
Binding ≤ 10μM
|
|
Z104296-1-O |
Cyclin-dependent Kinase 1/cyclin B1 (cluster #1 Of 2), Other |
Other |
160 |
0.29 |
Binding ≤ 10μM
|
|
Z104299-1-O |
Cyclin-dependent Kinase 2/cyclin E1 (cluster #1 Of 1), Other |
Other |
347 |
0.27 |
Binding ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
78 |
0.30 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
44 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
25 |
0.32 |
Functional ≤ 10μM
|
|
Z80942-1-O |
HEL (Embryonic Lung Cells) (cluster #1 Of 2), Other |
Other |
5 |
0.35 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80335-1-O |
NHDF (cluster #1 Of 2), Other |
Other |
73 |
0.30 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.49 |
7.93 |
-49.42 |
4 |
10 |
1 |
116 |
449.539 |
5 |
↓
|
|
|
|
Analogs
-
34583707
-
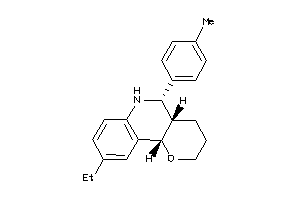
-
39283830
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
1190 |
0.55 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
2340 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
2.82 |
-9.44 |
2 |
4 |
0 |
75 |
204.181 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.06 |
-0.42 |
-14.67 |
2 |
6 |
0 |
83 |
232.243 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.06 |
-0.56 |
-57.78 |
3 |
6 |
1 |
84 |
233.251 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.27 |
Binding ≤ 10μM |
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
3330 |
0.20 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7020 |
0.19 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
940 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
2100 |
0.21 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3000 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.94 |
2.26 |
-55.93 |
1 |
6 |
1 |
59 |
596.977 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.29 |
Binding ≤ 10μM
|
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.27 |
Binding ≤ 10μM |
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
3330 |
0.20 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7020 |
0.19 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
940 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
2100 |
0.21 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3000 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.94 |
17.46 |
-55.26 |
1 |
6 |
1 |
60 |
596.977 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.24 |
Binding ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2480 |
0.26 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.33 |
Binding ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
280 |
0.31 |
Functional ≤ 10μM
|
|
Z80135-1-O |
F9 (cluster #1 Of 1), Other |
Other |
33 |
0.35 |
Functional ≤ 10μM
|
|
Z80140-1-O |
GBM (cluster #1 Of 1), Other |
Other |
550 |
0.29 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
490 |
0.29 |
Functional ≤ 10μM
|
|
Z80250-1-O |
Me665/2/21 (cluster #1 Of 1), Other |
Other |
420 |
0.30 |
Functional ≤ 10μM
|
|
Z80306-1-O |
NB-4 (Promyelocytic Leukemia Cells) (cluster #1 Of 1), Other |
Other |
400 |
0.30 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
130 |
0.32 |
Functional ≤ 10μM |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
430 |
0.30 |
Functional ≤ 10μM
|
|
Z80449-1-O |
SAOS-2 (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
670 |
0.29 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
440 |
0.30 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
510 |
0.29 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
680 |
0.29 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
430 |
0.30 |
Functional ≤ 10μM
|
|
Z81065-1-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
350 |
0.30 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
320 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.63 |
13.65 |
-51.5 |
1 |
3 |
-1 |
60 |
397.494 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
-
-
- API0007160
- API0007162
- API0007161
-
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
48 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
6.38 |
-44.83 |
3 |
6 |
1 |
68 |
410.538 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.98 |
4.93 |
-57.34 |
1 |
6 |
-1 |
70 |
408.522 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.98 |
6.26 |
-47.45 |
3 |
6 |
1 |
68 |
410.538 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.40 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z100501-1-O |
Lu1 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
80 |
0.52 |
Functional ≤ 10μM
|
|
Z103204-1-O |
A427 (cluster #1 Of 4), Other |
Other |
890 |
0.45 |
Functional ≤ 10μM
|
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
470 |
0.47 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
501 |
0.46 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
73 |
0.53 |
Functional ≤ 10μM
|
|
Z80008-2-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
890 |
0.45 |
Functional ≤ 10μM
|
|
Z80036-2-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #2 Of 3), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80145-1-O |
H69 (cluster #1 Of 4), Other |
Other |
1800 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-10-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #10 Of 12), Other |
Other |
660 |
0.46 |
Functional ≤ 10μM
|
|
Z80164-3-O |
HT-1080 (Fibrosarcoma Cells) (cluster #3 Of 6), Other |
Other |
4300 |
0.40 |
Functional ≤ 10μM
|
|
Z80186-10-O |
K562 (Erythroleukemia Cells) (cluster #10 Of 11), Other |
Other |
380 |
0.47 |
Functional ≤ 10μM
|
|
Z80193-4-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #4 Of 12), Other |
Other |
500 |
0.46 |
Functional ≤ 10μM |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
430 |
0.47 |
Functional ≤ 10μM
|
|
Z80322-1-O |
NCI-H358 (Lung Carcinama Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80509-1-O |
SNU-638 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.42 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
800 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
470 |
0.47 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
410 |
0.47 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
510 |
0.46 |
Functional ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 6), Other |
Other |
812 |
0.45 |
Functional ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 6), Other |
Other |
1780 |
0.42 |
Functional ≤ 10μM |
|
Z81170-5-O |
LNCaP (Prostate Carcinoma) (cluster #5 Of 5), Other |
Other |
3250 |
0.40 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
310 |
0.48 |
Functional ≤ 10μM
|
|
Z81252-10-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #10 Of 11), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z81277-1-O |
NCI-N417 (cluster #1 Of 2), Other |
Other |
870 |
0.45 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 4), Other |
Other |
2600 |
0.41 |
ADME/T ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 1), Other |
Other |
5010 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
190 |
0.50 |
ADME/T ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
3820 |
0.40 |
ADME/T ≤ 10μM |
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
5500 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
2070 |
0.42 |
ADME/T ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1820 |
0.42 |
ADME/T ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 1), Other |
Other |
3 |
0.63 |
ADME/T ≤ 10μM
|
|
Z81057-4-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #4 Of 5), Other |
Other |
370 |
0.47 |
ADME/T ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
5300 |
0.39 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
8.05 |
-7.84 |
1 |
2 |
0 |
29 |
246.313 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.28 |
8.52 |
-35.14 |
2 |
2 |
1 |
30 |
247.321 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
|
CLK3-1-E |
Dual Specificity Protein Kinase CLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
41 |
0.41 |
Binding ≤ 10μM
|
|
CSK21-1-E |
Casein Kinase II Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM
|
|
DAPK3-1-E |
Death-associated Protein Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.44 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
55 |
0.41 |
Binding ≤ 10μM
|
|
HIPK3-1-E |
Homeodomain-interacting Protein Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
45 |
0.41 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.41 |
Binding ≤ 10μM
|
|
TBK1-1-E |
Serine/threonine-protein Kinase TBK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
35 |
0.42 |
Binding ≤ 10μM
|
|
Z103203-2-O |
A375 (cluster #2 Of 3), Other |
Other |
3900 |
0.30 |
Functional ≤ 10μM
|
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
5300 |
0.30 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
8900 |
0.28 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
2100 |
0.32 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
3000 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
2200 |
0.32 |
Functional ≤ 10μM
|
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
4700 |
0.30 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
6400 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
10.5 |
-55.54 |
1 |
5 |
-1 |
78 |
348.769 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.84 |
5.03 |
-92.09 |
7 |
8 |
2 |
122 |
456.497 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
7.99 |
-52.68 |
3 |
7 |
1 |
83 |
362.498 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.19 |
6.32 |
-43.89 |
3 |
7 |
1 |
86 |
362.498 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.64 |
5.69 |
-104.44 |
8 |
9 |
2 |
142 |
511.577 |
6 |
↓
|
|
|
|
Analogs
-
4621972
-
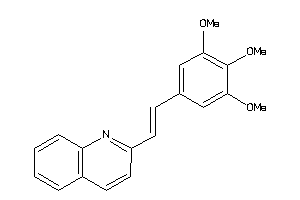
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.02 |
9.71 |
-12.42 |
0 |
4 |
0 |
41 |
321.376 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.02 |
9.74 |
-30.59 |
1 |
4 |
1 |
42 |
322.384 |
5 |
↓
|
|
|
|
Analogs
-
7586807
-
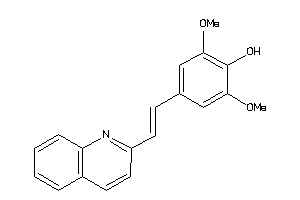
-
13137527
-
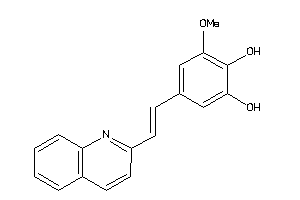
-
16980597
-
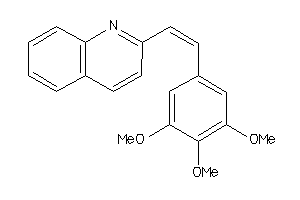
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.02 |
9.14 |
-12.54 |
0 |
4 |
0 |
41 |
321.376 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.02 |
9.45 |
-37.78 |
1 |
4 |
1 |
42 |
322.384 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
180 |
0.33 |
Binding ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
470 |
0.31 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
610 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
4.26 |
-10.59 |
3 |
7 |
0 |
100 |
385.379 |
2 |
↓
|
|
|
|
Analogs
-
34384993
-
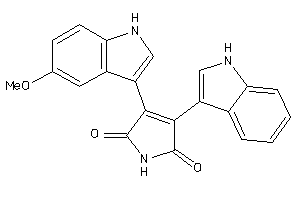
-
585159
-
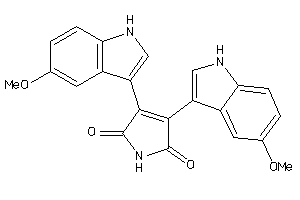
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
4640 |
0.26 |
Binding ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
300 |
0.31 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
1280 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
5.23 |
-13.49 |
3 |
7 |
0 |
100 |
387.395 |
4 |
↓
|
|
|
|
Analogs
-
32120145
-
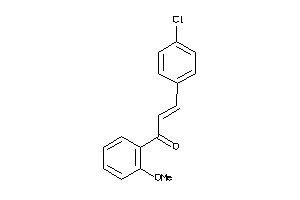
-
4100768
-
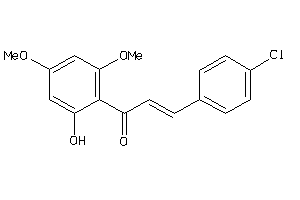
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
6500 |
0.35 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
6200 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
2.88 |
-11.47 |
0 |
3 |
0 |
35 |
302.757 |
5 |
↓
|
|
|
|
Analogs
-
3872981
-
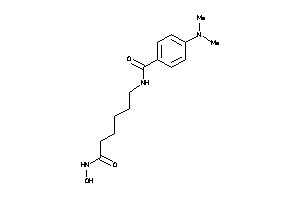
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
750 |
0.39 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
Z80532-3-O |
T-24 (Bladder Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
3800 |
0.34 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
800 |
0.39 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
2300 |
0.36 |
Functional ≤ 10μM
|
|
Z80877-1-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #1 Of 2), Other |
Other |
3900 |
0.34 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
210 |
0.42 |
Functional ≤ 10μM
|
|
Z81186-1-O |
LS174T (Colon Adencocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
1500 |
0.37 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
700 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
4.23 |
-22.59 |
3 |
6 |
0 |
82 |
307.394 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
2.26 |
2.24 |
-13.63 |
3 |
6 |
0 |
85 |
307.394 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.93 |
12.79 |
-11.95 |
2 |
8 |
0 |
86 |
432.528 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.93 |
13.07 |
-35.61 |
3 |
8 |
1 |
87 |
433.536 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.93 |
13.18 |
-31.89 |
3 |
8 |
1 |
87 |
433.536 |
9 |
↓
|
|
|
|
Analogs
-
602192
-
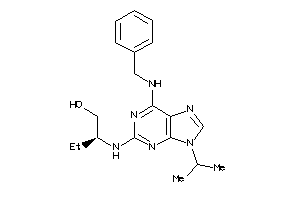
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.30 |
Binding ≤ 10μM
|
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CCNH-1-E |
Cyclin H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
540 |
0.34 |
Binding ≤ 10μM |
|
CCNT1-1-E |
Cyclin T1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
950 |
0.32 |
Binding ≤ 10μM |
|
CCNT2-1-E |
Cyclin-T2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
810 |
0.33 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM
|
|
CDK16-1-E |
Serine/threonine-protein Kinase PCTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
990 |
0.32 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.38 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.37 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.35 |
Binding ≤ 10μM
|
|
CDK7-1-E |
Cyclin-dependent Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.33 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
950 |
0.32 |
Binding ≤ 10μM |
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.31 |
Binding ≤ 10μM
|
|
CLK2-1-E |
Dual Specificity Protein Kinase CLK2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.33 |
Binding ≤ 10μM
|
|
CLK4-1-E |
Dual Specificity Protein Kinase CLK4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.29 |
Binding ≤ 10μM
|
|
DYR1A-3-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.29 |
Binding ≤ 10μM
|
|
DYR1B-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.32 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
260 |
0.35 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.35 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.30 |
Binding ≤ 10μM
|
|
KC1G2-1-E |
Casein Kinase I Gamma 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.32 |
Binding ≤ 10μM
|
|
KC1G3-1-E |
Casein Kinase I Isoform Gamma-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.30 |
Binding ≤ 10μM
|
|
KS6A2-1-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.30 |
Binding ≤ 10μM
|
|
MAT1-1-E |
CDK-activating Kinase Assembly Factor MAT1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
540 |
0.34 |
Binding ≤ 10μM |
|
PAK4-1-E |
Serine/threonine-protein Kinase PAK 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.28 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.31 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
280 |
0.35 |
Binding ≤ 10μM |
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
940 |
0.32 |
Binding ≤ 10μM
|
|
Z104296-2-O |
Cyclin-dependent Kinase 1/cyclin B1 (cluster #2 Of 2), Other |
Other |
450 |
0.34 |
Binding ≤ 10μM
|
|
Z104299-1-O |
Cyclin-dependent Kinase 2/cyclin E1 (cluster #1 Of 1), Other |
Other |
220 |
0.36 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
6310 |
0.28 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
6000 |
0.28 |
Functional ≤ 10μM
|
|
Z80390-10-O |
PC-3 (Prostate Carcinoma Cells) (cluster #10 Of 10), Other |
Other |
8000 |
0.27 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
5000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
9.65 |
-10.26 |
3 |
7 |
0 |
88 |
354.458 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.65 |
9.9 |
-28.83 |
4 |
7 |
1 |
89 |
355.466 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.65 |
9.47 |
-25.05 |
4 |
7 |
1 |
89 |
355.466 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
-2.96 |
-11.42 |
1 |
3 |
0 |
50 |
243.287 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-3-E |
Serine/threonine-protein Kinase Aurora-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
31 |
0.44 |
Binding ≤ 10μM |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
1600 |
0.34 |
Functional ≤ 10μM |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
7800 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
9.25 |
-15.42 |
2 |
6 |
0 |
76 |
317.352 |
3 |
↓
|
|
|
|
Analogs
-
11616249
-
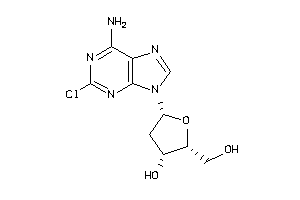
-
1545060
-
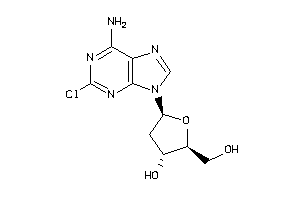
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100515-1-O |
HPAC (Pancreatic Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
9320 |
0.37 |
Functional ≤ 10μM |
|
Z100733-1-O |
CT26 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
131 |
0.51 |
Functional ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
1 |
0.66 |
Functional ≤ 10μM |
|
Z80132-1-O |
EL4 (Thymoma Cells) (cluster #1 Of 2), Other |
Other |
848 |
0.45 |
Functional ≤ 10μM |
|
Z80166-12-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #12 Of 12), Other |
Other |
9440 |
0.37 |
Functional ≤ 10μM |
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
7690 |
0.38 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
393 |
0.47 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
2350 |
0.41 |
Functional ≤ 10μM |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
8280 |
0.37 |
Functional ≤ 10μM |
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM |
|
Z80742-1-O |
C6 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
9070 |
0.37 |
Functional ≤ 10μM |
|
Z80928-2-O |
HCT-116 (Colon Carcinoma Cells) (cluster #2 Of 9), Other |
Other |
9430 |
0.37 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.10 |
-2.49 |
-11.31 |
4 |
8 |
0 |
119 |
285.691 |
2 |
↓
|
|
|
|
Analogs
-
4095791
-
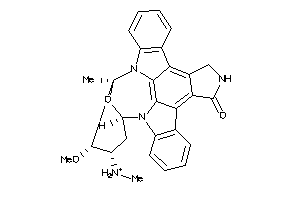
-
12501465
-
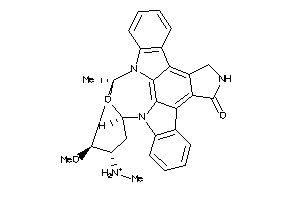
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAK1-1-E |
Adaptor-associated Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
AAPK1-1-E |
AMP-activated Protein Kinase, Alpha-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.32 |
Binding ≤ 10μM
|
|
AAPK2-1-E |
AMP-activated Protein Kinase, Alpha-2 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.32 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
91 |
0.28 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.27 |
Binding ≤ 10μM
|
|
ACK1-1-E |
Tyrosine Kinase Non-receptor Protein 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.31 |
Binding ≤ 10μM
|
|
ACV1B-1-E |
Activin Receptor Type-1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.25 |
Binding ≤ 10μM
|
|
ACVR1-1-E |
Activin Receptor Type-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.25 |
Binding ≤ 10μM
|
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
AKT2-2-E |
Serine/threonine-protein Kinase AKT2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.32 |
Binding ≤ 10μM
|
|
AKT3-2-E |
Serine/threonine-protein Kinase AKT3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM
|
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.30 |
Binding ≤ 10μM
|
|
ANKK1-1-E |
Ankyrin Repeat And Protein Kinase Domain-containing Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.26 |
Binding ≤ 10μM
|
|
AURKA-1-E |
Serine/threonine-protein Kinase Aurora-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
22 |
0.31 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.31 |
Binding ≤ 10μM
|
|
AURKC-1-E |
Serine/threonine-protein Kinase Aurora-C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.31 |
Binding ≤ 10μM
|
|
AVR2B-1-E |
Activin Receptor Type-2B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.21 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.31 |
Binding ≤ 10μM
|
|
BMP2K-1-E |
BMP-2-inducible Protein Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
BMR1A-1-E |
Bone Morphogenetic Protein Receptor Type-1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3700 |
0.22 |
Binding ≤ 10μM
|
|
BMX-1-E |
Tyrosine-protein Kinase BMX (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
410 |
0.26 |
Binding ≤ 10μM
|
|
BRSK1-1-E |
BR Serine/threonine-protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.30 |
Binding ≤ 10μM
|
|
BRSK2-1-E |
BR Serine/threonine-protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
BTK-1-E |
Tyrosine-protein Kinase BTK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.25 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM
|
|
CDK14-1-E |
Serine/threonine-protein Kinase PFTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.27 |
Binding ≤ 10μM
|
|
CDK16-1-E |
Serine/threonine-protein Kinase PCTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.30 |
Binding ≤ 10μM
|
|
CDK17-1-E |
Serine/threonine-protein Kinase PCTAIRE-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.28 |
Binding ≤ 10μM
|
|
CDK18-1-E |
Serine/threonine-protein Kinase PCTAIRE-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.26 |
Binding ≤ 10μM
|
|
CDK19-1-E |
Cell Division Cycle 2-like Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.27 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM
|
|
CDK3-1-E |
Cyclin-dependent Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.33 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.31 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
89 |
0.28 |
Binding ≤ 10μM
|
|
CDK6-1-E |
Cyclin-dependent Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
116 |
0.28 |
Binding ≤ 10μM
|
|
CDK7-1-E |
Cyclin-dependent Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
45 |
0.29 |
Binding ≤ 10μM
|
|
CDK8-1-E |
Cell Division Protein Kinase 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.25 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.32 |
Binding ≤ 10μM
|
|
CHK2-1-E |
Serine/threonine-protein Kinase Chk2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.33 |
Binding ≤ 10μM
|
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
32 |
0.30 |
Binding ≤ 10μM
|
|
CLK2-1-E |
Dual Specificity Protein Kinase CLK2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM
|
|
CLK3-1-E |
Dual Specificity Protein Kinase CLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
910 |
0.24 |
Binding ≤ 10μM
|
|
CLK4-1-E |
Dual Specificity Protein Kinase CLK4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.32 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.32 |
Binding ≤ 10μM
|
|
CSK-1-E |
Tyrosine-protein Kinase CSK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.25 |
Binding ≤ 10μM
|
|
CSK21-1-E |
Casein Kinase II Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7620 |
0.20 |
Binding ≤ 10μM
|
|
CSK22-1-E |
Casein Kinase II Alpha (prime) (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.33 |
Binding ≤ 10μM
|
|
CSK2B-1-E |
Casein Kinase II Beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.21 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.26 |
Binding ≤ 10μM
|
|
DAPK1-1-E |
Death-associated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
DAPK2-1-E |
Death-associated Protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
DAPK3-1-E |
Death-associated Protein Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.23 |
Binding ≤ 10μM
|
|
DCLK1-1-E |
Serine/threonine-protein Kinase DCLK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.28 |
Binding ≤ 10μM
|
|
DCLK2-1-E |
Serine/threonine-protein Kinase DCLK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
73 |
0.29 |
Binding ≤ 10μM
|
|
DCLK3-1-E |
Serine/threonine-protein Kinase DCLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.31 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.31 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
DMPK-1-E |
Myotonin-protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
DYR1B-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.30 |
Binding ≤ 10μM
|
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.28 |
Binding ≤ 10μM |
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
E2AK4-1-E |
Eukaryotic Translation Initiation Factor 2-alpha Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.31 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
96 |
0.28 |
Binding ≤ 10μM
|
|
EPHA1-1-E |
Ephrin Type-A Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.26 |
Binding ≤ 10μM
|
|
EPHA2-1-E |
Ephrin Type-A Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
870 |
0.24 |
Binding ≤ 10μM
|
|
EPHA3-1-E |
Ephrin Type-A Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
55 |
0.29 |
Binding ≤ 10μM
|
|
EPHA4-1-E |
Ephrin Type-A Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.25 |
Binding ≤ 10μM
|
|
EPHA5-1-E |
Ephrin Type-A Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
95 |
0.28 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.26 |
Binding ≤ 10μM
|
|
EPHA7-1-E |
Ephrin Type-A Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
690 |
0.25 |
Binding ≤ 10μM
|
|
EPHA8-1-E |
Ephrin Type-A Receptor 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.26 |
Binding ≤ 10μM
|
|
EPHB1-1-E |
Ephrin Type-B Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
590 |
0.25 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.23 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
233 |
0.27 |
Binding ≤ 10μM
|
|
ERBB4-1-E |
Receptor Protein-tyrosine Kinase ErbB-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
282 |
0.26 |
Binding ≤ 10μM
|
|
FAK1-1-E |
Focal Adhesion Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
68 |
0.29 |
Binding ≤ 10μM
|
|
FAK2-1-E |
Protein-tyrosine Kinase 2-beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
FER-1-E |
Tyrosine-protein Kinase FER (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.30 |
Binding ≤ 10μM
|
|
FES-1-E |
Tyrosine-protein Kinase FES (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.31 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.28 |
Binding ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.26 |
Binding ≤ 10μM
|
|
FGFR3-1-E |
Fibroblast Growth Factor Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
88 |
0.28 |
Binding ≤ 10μM
|
|
FGFR4-1-E |
Fibroblast Growth Factor Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.26 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.30 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
45 |
0.29 |
Binding ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
83 |
0.28 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
68 |
0.29 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
140 |
0.27 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
89 |
0.28 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
HIPK1-1-E |
Homeodomain-interacting Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2639 |
0.22 |
Binding ≤ 10μM
|
|
IGF1R-1-E |
Insulin-like Growth Factor 1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.30 |
Binding ≤ 10μM
|
|
IKKA-2-E |
Inhibitor Of Nuclear Factor Kappa B Kinase Alpha Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
39 |
0.30 |
Binding ≤ 10μM
|
|
IKKB-1-E |
Inhibitor Of Nuclear Factor Kappa B Kinase Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
771 |
0.24 |
Binding ≤ 10μM
|
|
IKKE-1-E |
Inhibitor Of Nuclear Factor Kappa B Kinase Epsilon Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM
|
|
INSR-1-E |
Insulin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.27 |
Binding ≤ 10μM
|
|
INSRR-1-E |
Insulin Receptor-related Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
81 |
0.28 |
Binding ≤ 10μM
|
|
IRAK3-1-E |
Interleukin-1 Receptor-associated Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
13 |
0.32 |
Binding ≤ 10μM
|
|
IRAK4-1-E |
Interleukin-1 Receptor-associated Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
34 |
0.30 |
Binding ≤ 10μM
|
|
ITK-1-E |
Tyrosine-protein Kinase ITK/TSK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.31 |
Binding ≤ 10μM
|
|
JAK2-1-E |
Tyrosine-protein Kinase JAK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM
|
|
JAK3-1-E |
Tyrosine-protein Kinase JAK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
KAP2-1-E |
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.28 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.30 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.28 |
Binding ≤ 10μM
|
|
KC1A-1-E |
Casein Kinase I Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.23 |
Binding ≤ 10μM
|
|
KC1AL-1-E |
Casein Kinase I Isoform Alpha-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.26 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.23 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.23 |
Binding ≤ 10μM
|
|
KC1G1-1-E |
Casein Kinase I Gamma 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.23 |
Binding ≤ 10μM
|
|
KC1G2-1-E |
Casein Kinase I Gamma 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.24 |
Binding ≤ 10μM
|
|
KCC1A-1-E |
CaM Kinase I Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
|
KCC1D-1-E |
CaM Kinase I Delta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.35 |
Binding ≤ 10μM
|
|
KCC1G-1-E |
CaM Kinase I Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.29 |
Binding ≤ 10μM
|
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
KCC2D-1-E |
CaM Kinase II Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
KCC4-1-E |
CaM Kinase IV (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
41 |
0.30 |
Binding ≤ 10μM
|
|
KGP1-1-E |
CGMP-dependent Protein Kinase 1 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.30 |
Binding ≤ 10μM
|
|
KGP2-1-E |
CGMP-dependent Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.31 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.32 |
Binding ≤ 10μM
|
|
KKCC1-1-E |
CaM-kinase Kinase Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
KKCC2-1-E |
CaM-kinase Kinase Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
KPB1-1-E |
Phosphorylase Kinase Alpha M Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
KPB2-1-E |
Phosphorylase Kinase Alpha L Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
KPBB-1-E |
Phosphorylase Kinase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM |
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.32 |
Binding ≤ 10μM |
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM |
|
KPCD1-1-E |
Protein Kinase C Mu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM |
|
KPCD2-1-E |
Serine/threonine-protein Kinase D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
72 |
0.29 |
Binding ≤ 10μM
|
|
KPCD3-1-E |
Protein Kinase C Nu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM |
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM |
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.32 |
Binding ≤ 10μM |
|
KPCI-1-E |
Protein Kinase C Iota (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM |
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.32 |
Binding ≤ 10μM |
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.32 |
Binding ≤ 10μM |
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM |
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM |
|
KS6A2-1-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.32 |
Binding ≤ 10μM
|
|
KS6A3-1-E |
Ribosomal Protein S6 Kinase Alpha 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
69 |
0.29 |
Binding ≤ 10μM
|
|
KS6A4-1-E |
Ribosomal Protein S6 Kinase Alpha 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM |
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
890 |
0.24 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM |
|
KS6B1-1-E |
Ribosomal Protein S6 Kinase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM |
|
KS6B2-1-E |
Ribosomal Protein S6 Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM |
|
KSYK-1-E |
Tyrosine-protein Kinase SYK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.33 |
Binding ≤ 10μM
|
|
LATS1-1-E |
Serine/threonine-protein Kinase LATS1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
LATS2-1-E |
Serine/threonine-protein Kinase LATS2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.31 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
LIMK1-1-E |
LIM Domain Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
690 |
0.25 |
Binding ≤ 10μM
|
|
LIMK2-1-E |
LIM Domain Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9200 |
0.20 |
Binding ≤ 10μM
|
|
LTK-1-E |
Leukocyte Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.30 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
|
M3K10-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
52 |
0.29 |
Binding ≤ 10μM
|
|
M3K11-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
M3K14-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 14 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.23 |
Binding ≤ 10μM
|
|
M3K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.23 |
Binding ≤ 10μM
|
|
M3K5-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.30 |
Binding ≤ 10μM
|
|
M3K7-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
35 |
0.30 |
Binding ≤ 10μM
|
|
M3K9-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 9 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.32 |
Binding ≤ 10μM
|
|
M4K1-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
M4K3-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM
|
|
M4K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.30 |
Binding ≤ 10μM
|
|
M4K5-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.30 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
880 |
0.24 |
Binding ≤ 10μM
|
|
MARK1-1-E |
Serine/threonine-protein Kinase MARK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
MARK2-1-E |
MAP/microtubule Affinity-regulating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
MARK3-1-E |
Serine/threonine-protein Kinase C-TAK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
MARK4-1-E |
MAP/microtubule Affinity-regulating Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM
|
|
MELK-1-E |
Maternal Embryonic Leucine Zipper Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.30 |
Binding ≤ 10μM
|
|
MERTK-1-E |
Proto-oncogene Tyrosine-protein Kinase MER (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.27 |
Binding ≤ 10μM
|
|
MINK1-1-E |
Misshapen-like Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7300 |
0.21 |
Binding ≤ 10μM
|
|
MK03-1-E |
MAP Kinase ERK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8400 |
0.20 |
Binding ≤ 10μM
|
|
MK07-1-E |
Mitogen-activated Protein Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.23 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
520 |
0.25 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.24 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
340 |
0.26 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.24 |
Binding ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
92 |
0.28 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.24 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.24 |
Binding ≤ 10μM
|
|
MK15-1-E |
Mitogen-activated Protein Kinase 15 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
35 |
0.30 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.27 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.31 |
Binding ≤ 10μM
|
|
MP2K1-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
670 |
0.25 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.30 |
Binding ≤ 10μM
|
|
MP2K3-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM
|
|
MP2K4-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.29 |
Binding ≤ 10μM
|
|
MP2K6-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
57 |
0.29 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
42 |
0.30 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.30 |
Binding ≤ 10μM
|
|
MST4-1-E |
Serine/threonine-protein Kinase MST4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.27 |
Binding ≤ 10μM
|
|
MUSK-1-E |
Muscle, Skeletal Receptor Tyrosine Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
86 |
0.28 |
Binding ≤ 10μM
|
|
MYLK-1-E |
Myosin Light Chain Kinase, Smooth Muscle (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
|
MYLK2-1-E |
Myosin Light Chain Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
61 |
0.29 |
Binding ≤ 10μM
|
|
MYLK4-1-E |
Myosin Light Chain Kinase Family Member 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.25 |
Binding ≤ 10μM
|
|
MYO3A-1-E |
Myosin IIIA (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.25 |
Binding ≤ 10μM
|
|
MYO3B-1-E |
Myosin-IIIB (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.26 |
Binding ≤ 10μM
|
|
NEK1-1-E |
Serine/threonine-protein Kinase Nek1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.24 |
Binding ≤ 10μM
|
|
NEK2-1-E |
Serine/threonine-protein Kinase NEK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
750 |
0.24 |
Binding ≤ 10μM
|
|
NEK5-1-E |
Serine/threonine-protein Kinase Nek5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
810 |
0.24 |
Binding ≤ 10μM
|
|
NEK7-1-E |
Serine/threonine-protein Kinase NEK7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.21 |
Binding ≤ 10μM
|
|
NEK9-1-E |
Serine/threonine-protein Kinase NEK9 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6700 |
0.21 |
Binding ≤ 10μM
|
|
NLK-1-E |
Serine/threonine Protein Kinase NLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.27 |
Binding ≤ 10μM
|
|
NTRK1-1-E |
Nerve Growth Factor Receptor Trk-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
NTRK2-1-E |
Neurotrophic Tyrosine Kinase Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
NTRK3-1-E |
NT-3 Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.31 |
Binding ≤ 10μM
|
|
NUAK1-1-E |
NUAK Family SNF1-like Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
NUAK2-1-E |
NUAK Family SNF1-like Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
P90584-2-E |
Protein Kinase Pfmrk (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.22 |
Binding ≤ 10μM
|
|
PAK1-1-E |
Serine/threonine-protein Kinase PAK 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.35 |
Binding ≤ 10μM
|
|
PAK2-1-E |
Serine/threonine-protein Kinase PAK 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
PAK3-1-E |
Serine/threonine-protein Kinase PAK 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.33 |
Binding ≤ 10μM
|
|
PAK4-1-E |
Serine/threonine-protein Kinase PAK 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM
|
|
PAK6-1-E |
Serine/threonine-protein Kinase PAK6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
PAK7-1-E |
Serine/threonine-protein Kinase PAK7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
PDPK1-1-E |
3-phosphoinositide Dependent Protein Kinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.35 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.35 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
PHKG1-1-E |
Phosphorylase Kinase Gamma Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM |
|
PHKG2-1-E |
Phosphorylase Kinase Gamma Subunit 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM |
|
PI42B-1-E |
Phosphatidylinositol-5-phosphate 4-kinase Type-2 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Binding ≤ 10μM
|
|
PI51A-1-E |
Phosphatidylinositol-4-phosphate 5-kinase Type-1 Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.32 |
Binding ≤ 10μM
|
|
PIM2-1-E |
Serine/threonine-protein Kinase PIM2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.33 |
Binding ≤ 10μM
|
|
PIM3-1-E |
Serine/threonine-protein Kinase PIM3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
PKN1-1-E |
Protein Kinase N1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
PKN2-1-E |
Protein Kinase N2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
PLK1-1-E |
Serine/threonine-protein Kinase PLK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.27 |
Binding ≤ 10μM
|
|
PLK2-1-E |
Serine/threonine-protein Kinase PLK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
197 |
0.27 |
Binding ≤ 10μM
|
|
PLK3-1-E |
Serine/threonine-protein Kinase PLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
910 |
0.24 |
Binding ≤ 10μM
|
|
PLK4-1-E |
Serine/threonine-protein Kinase PLK4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
PRKX-1-E |
Serine/threonine-protein Kinase PRKX (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
13 |
0.32 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.23 |
Binding ≤ 10μM
|
|
Q95J97-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.28 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.30 |
Binding ≤ 10μM
|
|
RIOK1-1-E |
Serine/threonine-protein Kinase RIO1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
130 |
0.28 |
Binding ≤ 10μM
|
|
RIOK3-1-E |
Serine/threonine-protein Kinase RIO3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
76 |
0.28 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5200 |
0.21 |
Binding ≤ 10μM
|
|
ROCK1-1-E |
Rho-associated Protein Kinase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
ROS1-1-E |
Proto-oncogene Tyrosine-protein Kinase ROS (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.30 |
Binding ≤ 10μM
|
|
SIK1-1-E |
Serine/threonine-protein Kinase SIK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.35 |
Binding ≤ 10μM
|
|
SIK2-1-E |
Serine/threonine-protein Kinase SIK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
86 |
0.28 |
Binding ≤ 10μM
|
|
SRMS-1-E |
Tyrosine-protein Kinase Srms (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.25 |
Binding ≤ 10μM
|
|
SRPK1-1-E |
Serine/threonine-protein Kinase SRPK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
SRPK2-1-E |
Serine/threonine-protein Kinase SRPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
67 |
0.29 |
Binding ≤ 10μM
|
|
ST17A-1-E |
Serine/threonine-protein Kinase 17A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.31 |
Binding ≤ 10μM
|
|
ST17B-1-E |
Serine/threonine-protein Kinase 17B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.30 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
98 |
0.28 |
Binding ≤ 10μM
|
|
ST32C-1-E |
Serine/threonine-protein Kinase 32C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.26 |
Binding ≤ 10μM
|
|
ST38L-1-E |
Serine/threonine-protein Kinase 38-like (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
STK11-1-E |
Serine/threonine-protein Kinase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
61 |
0.29 |
Binding ≤ 10μM
|
|
STK16-1-E |
Serine/threonine-protein Kinase 16 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.26 |
Binding ≤ 10μM
|
|
STK24-1-E |
Serine/threonine-protein Kinase 24 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.28 |
Binding ≤ 10μM
|
|
STK25-1-E |
Serine/threonine-protein Kinase 25 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.28 |
Binding ≤ 10μM
|
|
STK3-1-E |
Serine/threonine-protein Kinase MST2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
STK33-1-E |
Serine/threonine-protein Kinase 33 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.35 |
Binding ≤ 10μM
|
|
STK36-1-E |
Serine/threonine-protein Kinase 36 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6700 |
0.21 |
Binding ≤ 10μM
|
|
STK4-1-E |
Serine/threonine-protein Kinase MST1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
SYK-1-E |
Lysyl-tRNA Synthetase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.32 |
Binding ≤ 10μM
|
|
TAB1-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 7-interacting Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
35 |
0.30 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
770 |
0.24 |
Binding ≤ 10μM
|
|
TEC-1-E |
Tyrosine-protein Kinase TEC (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.24 |
Binding ≤ 10μM
|
|
TESK1-1-E |
Dual Specificity Testis-specific Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.26 |
Binding ≤ 10μM
|
|
TGFR1-1-E |
TGF-beta Receptor Type I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Binding ≤ 10μM
|
|
TIE1-1-E |
Tyrosine-protein Kinase Receptor Tie-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.29 |
Binding ≤ 10μM
|
|
TIE2-1-E |
Tyrosine-protein Kinase TIE-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
52 |
0.29 |
Binding ≤ 10μM
|
|
TLK1-1-E |
Serine/threonine-protein Kinase Tousled-like 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.30 |
Binding ≤ 10μM
|
|
TLK2-1-E |
Serine/threonine-protein Kinase Tousled-like 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.31 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.24 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
TNK1-1-E |
Non-receptor Tyrosine-protein Kinase TNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
TSSK1-1-E |
Testis-specific Serine/threonine-protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.30 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
61 |
0.29 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.27 |
Binding ≤ 10μM
|
|
TYK2-1-E |
Tyrosine-protein Kinase TYK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.29 |
Binding ≤ 10μM
|
|
TYRO3-1-E |
Tyrosine-protein Kinase Receptor TYRO3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
49 |
0.29 |
Binding ≤ 10μM
|
|
UFO-1-E |
Tyrosine-protein Kinase Receptor UFO (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.33 |
Binding ≤ 10μM
|
|
ULK3-1-E |
Serine/threonine-protein Kinase ULK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
14 |
0.31 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
88 |
0.28 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.31 |
Binding ≤ 10μM
|
|
WEE1-1-E |
Serine/threonine-protein Kinase WEE1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.23 |
Binding ≤ 10μM
|
|
YES-1-E |
Tyrosine-protein Kinase YES (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
52 |
0.29 |
Binding ≤ 10μM
|
|
ZAP70-1-E |
Tyrosine-protein Kinase ZAP-70 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
53 |
0.29 |
Binding ≤ 10μM
|
|
AKT1-1-E |
Serine/threonine-protein Kinase AKT (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.25 |
Functional ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Functional ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
10 |
0.32 |
Binding ≤ 10μM
|
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
59 |
0.29 |
Binding ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
16 |
0.31 |
Functional ≤ 10μM
|
|
Z100201-1-O |
Splenocytes (Spleen Cells) (cluster #1 Of 1), Other |
Other |
400 |
0.26 |
Functional ≤ 10μM |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
190 |
0.27 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
7500 |
0.20 |
Functional ≤ 10μM |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
9 |
0.32 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
3 |
0.34 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
1 |
0.36 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
50 |
0.29 |
Functional ≤ 10μM
|
|
Z80800-1-O |
MIA PaCa-2 (Pancreatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
370 |
0.26 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
6 |
0.33 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
77 |
0.28 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
4 |
0.34 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
71 |
0.29 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
8.49 |
-39.52 |
3 |
7 |
1 |
74 |
467.549 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.89 |
8.85 |
-16.32 |
2 |
7 |
0 |
69 |
466.541 |
2 |
↓
|
|
|
|
Analogs
-
2168378
-
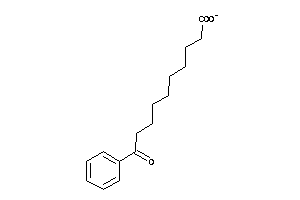
-
2378750
-
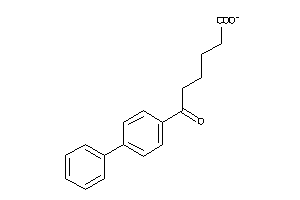
-
2378751
-
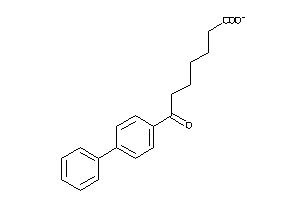
-
2579890
-
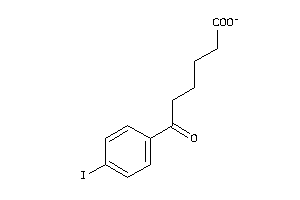
-
2579891
-
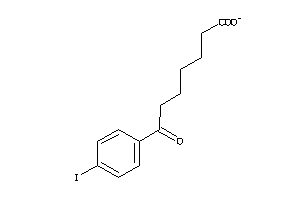
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDA11-2-E |
Histone Deacetylase 11 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC2-3-E |
Histone Deacetylase 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC3-3-E |
Histone Deacetylase 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
Z80928-4-O |
HCT-116 (Colon Carcinoma Cells) (cluster #4 Of 9), Other |
Other |
1720 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
1.66 |
-50.41 |
0 |
3 |
-1 |
57 |
233.287 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.73 |
12.27 |
-52.74 |
1 |
3 |
-1 |
60 |
455.703 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.73 |
10.3 |
-5.37 |
2 |
3 |
0 |
58 |
456.711 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.21 |
9.9 |
-24.79 |
3 |
5 |
1 |
64 |
297.382 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.21 |
9.53 |
-8.73 |
2 |
5 |
0 |
63 |
296.374 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.30 |
5.16 |
-16.72 |
2 |
10 |
0 |
127 |
377.43 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 6), Bacterial |
Bacteria |
8000 |
0.30 |
Binding ≤ 10μM
|
|
AMPH-1-B |
Penicillin-binding Protein AmpH (cluster #1 Of 2), Bacterial |
Bacteria |
8000 |
0.30 |
Binding ≤ 10μM
|
|
MDH-1-B |
Malate Dehydrogenase (cluster #1 Of 1), Bacterial |
Bacteria |
8000 |
0.30 |
Binding ≤ 10μM
|
|
CTRB1-3-E |
Beta-chymotrypsin (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.29 |
Binding ≤ 10μM
|
|
GLI1-1-E |
Zinc Finger Protein GLI1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.34 |
Binding ≤ 10μM
|
|
GLI2-1-E |
Zinc Finger Protein GLI2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.32 |
Binding ≤ 10μM
|
|
JAK3-2-E |
Tyrosine-protein Kinase JAK3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
61 |
0.42 |
Binding ≤ 10μM
|
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
184 |
0.39 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
184 |
0.39 |
Binding ≤ 10μM
|
|
KCC2D-1-E |
CaM Kinase II Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
184 |
0.39 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
184 |
0.39 |
Binding ≤ 10μM
|
|
KGP1-1-E |
CGMP-dependent Protein Kinase 1 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
650 |
0.36 |
Binding ≤ 10μM |
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
680 |
0.36 |
Binding ≤ 10μM |
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
680 |
0.36 |
Binding ≤ 10μM |
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
680 |
0.36 |
Binding ≤ 10μM |
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
680 |
0.36 |
Binding ≤ 10μM |
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
M3K11-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
49 |
0.43 |
Binding ≤ 10μM
|
|
M3K9-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 9 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
138 |
0.40 |
Binding ≤ 10μM
|
|
MYLK-1-E |
Myosin Light Chain Kinase, Smooth Muscle (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.32 |
Binding ≤ 10μM |
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
830 |
0.35 |
Binding ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
4800 |
0.31 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
4000 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
1720 |
0.34 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
3200 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
6.35 |
-15.15 |
3 |
4 |
0 |
61 |
311.344 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
3.27 |
-16.33 |
1 |
4 |
0 |
49 |
434.483 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
2.34 |
-17.94 |
1 |
4 |
0 |
49 |
434.483 |
6 |
↓
|
|
|
|
Analogs
-
12492969
-
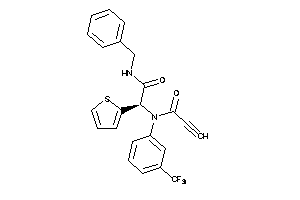
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
2.71 |
-16.93 |
1 |
4 |
0 |
49 |
442.462 |
7 |
↓
|
|
|
|
Analogs
-
12492964
-
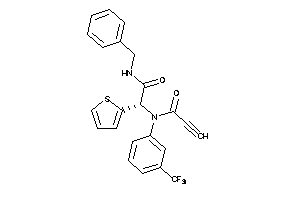
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
2.64 |
-16.68 |
1 |
4 |
0 |
49 |
442.462 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATR-1-E |
Serine-protein Kinase ATR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
1100 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
6.41 |
-46.56 |
4 |
8 |
1 |
119 |
440.549 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.90 |
4.83 |
-15 |
3 |
8 |
0 |
118 |
439.541 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATR-1-E |
Serine-protein Kinase ATR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
1100 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
6.27 |
-47.57 |
4 |
8 |
1 |
119 |
440.549 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.90 |
4.18 |
-15.12 |
3 |
8 |
0 |
118 |
439.541 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80186-5-O |
K562 (Erythroleukemia Cells) (cluster #5 Of 11), Other |
Other |
96 |
0.49 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
120 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
6.67 |
-12.94 |
1 |
4 |
0 |
60 |
268.268 |
2 |
↓
|
|
|
|
Analogs
-
9300392
-
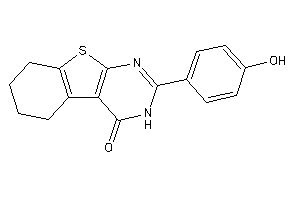
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
7.64 |
-11.7 |
1 |
4 |
0 |
55 |
312.394 |
2 |
↓
|
|
|
|
Analogs
-
3917708
-
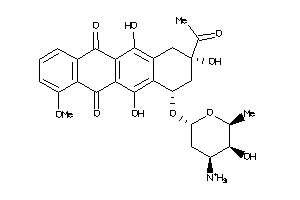
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-1-E |
Serine/threonine-protein Kinase Aurora-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.27 |
Binding ≤ 10μM |
|
MMP2-2-E |
72 KDa Type IV Collagenase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.21 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.22 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 2), Other |
Other |
2600 |
0.20 |
Binding ≤ 10μM
|
|
Z103196-1-O |
22RV1 (cluster #1 Of 2), Other |
Other |
1300 |
0.21 |
Functional ≤ 10μM |
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
130 |
0.25 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
370 |
0.23 |
Functional ≤ 10μM
|
|
Z103219-1-O |
Ca9-22 (cluster #1 Of 1), Other |
Other |
1300 |
0.21 |
Functional ≤ 10μM
|
|
Z50347-3-O |
Saccharomyces Cerevisiae (cluster #3 Of 3), Other |
Other |
10000 |
0.18 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z80008-1-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80017-1-O |
A2780cisR (Cisplatin-resistant Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
6 |
0.30 |
Functional ≤ 10μM
|
|
Z80018-2-O |
A-375 (Malignant Melanoma Cells) (cluster #2 Of 4), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM |
|
Z80021-3-O |
A498 (Renal Carcinoma Cells) (cluster #3 Of 5), Other |
Other |
53 |
0.26 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
700 |
0.22 |
Functional ≤ 10μM |
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
310 |
0.23 |
Functional ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
8 |
0.29 |
Functional ≤ 10μM
|
|
Z80039-1-O |
BGC-823 (Stomach Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
1000 |
0.22 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80068-1-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
6000 |
0.19 |
Functional ≤ 10μM
|
|
Z80092-1-O |
CHO-K1 (Ovarian Cells) (cluster #1 Of 2), Other |
Other |
400 |
0.23 |
Functional ≤ 10μM
|
|
Z80105-2-O |
COR-L23 (Lung Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
319 |
0.23 |
Functional ≤ 10μM
|
|
Z80114-1-O |
Daudi (Burkitts Lymphoma Cells) (cluster #1 Of 4), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
2540 |
0.20 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
6240 |
0.19 |
Functional ≤ 10μM |
|
Z80126-2-O |
DuPro (cluster #2 Of 2), Other |
Other |
1800 |
0.21 |
Functional ≤ 10μM
|
|
Z80138-1-O |
G-361 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
870 |
0.22 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
920 |
0.22 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
940 |
0.22 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
4700 |
0.19 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
9320 |
0.18 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
92 |
0.25 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
950 |
0.22 |
Functional ≤ 10μM
|
|
Z80175-1-O |
IMR-32 (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
430 |
0.23 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
21 |
0.28 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
900 |
0.22 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
200 |
0.24 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
9700 |
0.18 |
Functional ≤ 10μM
|
|
Z80214-2-O |
M14 (Melanoma Cells) (cluster #2 Of 3), Other |
Other |
820 |
0.22 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
80 |
0.25 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
920 |
0.22 |
Functional ≤ 10μM
|
|
Z80227-1-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM
|
|
Z80240-1-O |
MDA-MB-361 (cluster #1 Of 1), Other |
Other |
33 |
0.27 |
Functional ≤ 10μM
|
|
Z80243-1-O |
MDA-MB-453 (Breast Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
120 |
0.25 |
Functional ≤ 10μM
|
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
7 |
0.29 |
Functional ≤ 10μM
|
|
Z80254-1-O |
MES-SA (Uterine Sarcoma Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
704 |
0.22 |
Functional ≤ 10μM
|
|
Z80267-1-O |
MKN-1 (cluster #1 Of 1), Other |
Other |
640 |
0.22 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
3130 |
0.20 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z80295-6-O |
MT4 (Lymphocytes) (cluster #6 Of 8), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80322-2-O |
NCI-H358 (Lung Carcinama Cells) (cluster #2 Of 2), Other |
Other |
620 |
0.22 |
Functional ≤ 10μM
|
|
Z80330-1-O |
NCI-N87 (cluster #1 Of 1), Other |
Other |
420 |
0.23 |
Functional ≤ 10μM
|
|
Z80354-4-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
35 |
0.27 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
17 |
0.28 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
830 |
0.22 |
Functional ≤ 10μM |
|
Z80364-1-O |
P388/ADR (Lymphoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80367-1-O |
P388/S (cluster #1 Of 1), Other |
Other |
13 |
0.28 |
Functional ≤ 10μM
|
|
Z80389-1-O |
PC-14 (cluster #1 Of 1), Other |
Other |
290 |
0.23 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
680 |
0.22 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
912 |
0.22 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
560 |
0.22 |
Functional ≤ 10μM
|
|
Z80441-1-O |
RXF 944 (cluster #1 Of 1), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
600 |
0.22 |
Functional ≤ 10μM |
|
Z80468-1-O |
SF-295 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
35 |
0.27 |
Functional ≤ 10μM
|
|
Z80472-1-O |
SiHa (Cervical Squamous Cell Carcinoma) (cluster #1 Of 2), Other |
Other |
500 |
0.23 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80477-1-O |
SK_HEP1 (Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z80479-1-O |
SK-MEL (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
1730 |
0.21 |
Functional ≤ 10μM
|
|
Z80481-1-O |
SK-MEL-1 (Metastatic Melanoma Cells) (cluster #1 Of 1), Other |
Other |
3092 |
0.20 |
Functional ≤ 10μM |
|
Z80482-3-O |
SK-MEL-2 (Melanoma Cells) (cluster #3 Of 4), Other |
Other |
97 |
0.25 |
Functional ≤ 10μM
|
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
600 |
0.22 |
Functional ≤ 10μM |
|
Z80490-3-O |
SK-MES-1 (cluster #3 Of 4), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80491-2-O |
SK-N-MC (Neuroepithelioma Cells) (cluster #2 Of 4), Other |
Other |
120 |
0.25 |
Functional ≤ 10μM
|
|
Z80492-1-O |
SK-N-SH (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
1100 |
0.21 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
800 |
0.22 |
Functional ≤ 10μM
|
|
Z80495-1-O |
SK-VLB (cluster #1 Of 1), Other |
Other |
6540 |
0.19 |
Functional ≤ 10μM
|
|
Z80503-1-O |
SNU1 (cluster #1 Of 2), Other |
Other |
4000 |
0.19 |
Functional ≤ 10μM
|
|
Z80509-1-O |
SNU-638 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
690 |
0.22 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
640 |
0.22 |
Functional ≤ 10μM
|
|
Z80559-1-O |
U251 (cluster #1 Of 3), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80560-1-O |
U-251 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80574-2-O |
UMUC3 (cluster #2 Of 2), Other |
Other |
9 |
0.29 |
Functional ≤ 10μM
|
|
Z80581-1-O |
VA13 (cluster #1 Of 1), Other |
Other |
400 |
0.23 |
Functional ≤ 10μM
|
|
Z80581-1-O |
VA13 (cluster #1 Of 1), Other |
Other |
699 |
0.22 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
5000 |
0.19 |
Functional ≤ 10μM
|
|
Z80595-1-O |
WIL2-NS (Lymphoblastoid Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.24 |
Functional ≤ 10μM |
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
44 |
0.26 |
Functional ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
9 |
0.29 |
Functional ≤ 10μM
|
|
Z80640-1-O |
786-0 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
3100 |
0.20 |
Functional ≤ 10μM |
|
Z80661-1-O |
A704 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.20 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
540 |
0.22 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
970 |
0.22 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
1000 |
0.22 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
72 |
0.26 |
Functional ≤ 10μM
|
|
Z80726-1-O |
TRAMP-C1A (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
76 |
0.26 |
Functional ≤ 10μM
|
|
Z80730-1-O |
C2D Cell Line (cluster #1 Of 1), Other |
Other |
47 |
0.26 |
Functional ≤ 10μM
|
|
Z80731-1-O |
C2G Cell Line (cluster #1 Of 1), Other |
Other |
29 |
0.27 |
Functional ≤ 10μM
|
|
Z80732-1-O |
TRAMP-C2H (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80735-1-O |
C38 (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80768-1-O |
CH1 (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
6 |
0.30 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
520 |
0.23 |
Functional ≤ 10μM
|
|
Z80801-2-O |
CRL-7065 Cell Line (cluster #2 Of 2), Other |
Other |
500 |
0.23 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
368 |
0.23 |
Functional ≤ 10μM |
|
Z80846-1-O |
F460pv8/eto Cell Line (cluster #1 Of 2), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80887-1-O |
H2981 Cell Line (cluster #1 Of 1), Other |
Other |
700 |
0.22 |
Functional ≤ 10μM
|
|
Z80889-1-O |
NCI-H322M (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
27 |
0.27 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
6810 |
0.19 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
900 |
0.22 |
Functional ≤ 10μM |
|
Z80943-1-O |
HEL 299 (cluster #1 Of 1), Other |
Other |
1700 |
0.21 |
Functional ≤ 10μM |
|
Z80945-1-O |
HEp-2 (Laryngeal Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
59 |
0.26 |
Functional ≤ 10μM
|
|
Z81005-1-O |
HTB-54 Cell Line (cluster #1 Of 1), Other |
Other |
1100 |
0.21 |
Functional ≤ 10μM
|
|
Z81006-1-O |
Human Breast 401NL Tumor Cell Line (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
620 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
954 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
1290 |
0.21 |
Functional ≤ 10μM |
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
660 |
0.22 |
Functional ≤ 10μM
|
|
Z81025-1-O |
Human Lung LXF 529L Tumor Cell Line (cluster #1 Of 1), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z81026-1-O |
Human Lung LXF 629L Tumor Cell Line (cluster #1 Of 1), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM
|
|
Z81028-1-O |
Human Melanoma Cell Line (cluster #1 Of 1), Other |
Other |
280 |
0.24 |
Functional ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z81043-1-O |
N592 (cluster #1 Of 2), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z81101-1-O |
KBM-3 Cell Line (cluster #1 Of 1), Other |
Other |
26 |
0.27 |
Functional ≤ 10μM
|
|
Z81102-1-O |
KBM-3/DOX Cell Line (cluster #1 Of 1), Other |
Other |
490 |
0.23 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
600 |
0.22 |
Functional ≤ 10μM |
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
7010 |
0.19 |
Functional ≤ 10μM
|
|
Z81133-1-O |
L1210 (1565) (cluster #1 Of 1), Other |
Other |
315 |
0.23 |
Functional ≤ 10μM |
|
Z81134-1-O |
L2987 (Lung Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81164-1-O |
LL Cell Line (cluster #1 Of 2), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81167-1-O |
LLTC Cell Line (cluster #1 Of 1), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
78 |
0.26 |
Functional ≤ 10μM
|
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
42 |
0.26 |
Functional ≤ 10μM
|
|
Z81201-1-O |
HOP-62 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
4 |
0.30 |
Functional ≤ 10μM
|
|
Z81211-1-O |
LXFL529 Cell Line (cluster #1 Of 1), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
79 |
0.25 |
Functional ≤ 10μM |
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
960 |
0.22 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
710 |
0.22 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
730 |
0.22 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
840 |
0.22 |
Functional ≤ 10μM
|
|
Z81257-1-O |
MEL-AC Cell Line (cluster #1 Of 1), Other |
Other |
1200 |
0.21 |
Functional ≤ 10μM
|
|
Z81261-1-O |
UACC-375 (cluster #1 Of 1), Other |
Other |
112 |
0.25 |
Functional ≤ 10μM
|
|
Z81277-2-O |
NCI-N417 (cluster #2 Of 2), Other |
Other |
230 |
0.24 |
Functional ≤ 10μM
|
|
Z81280-1-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
8 |
0.29 |
Functional ≤ 10μM
|
|
Z81284-1-O |
NCI-H647 (cluster #1 Of 2), Other |
Other |
1 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
80 |
0.25 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
820 |
0.22 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
2320 |
0.20 |
Functional ≤ 10μM
|
|
Z81341-1-O |
Tumor Cell Lines (cluster #1 Of 1), Other |
Other |
74 |
0.26 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 2), Other |
Other |
500 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
100 |
0.25 |
ADME/T ≤ 10μM
|
|
Z80005-1-O |
3T3 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
3700 |
0.19 |
ADME/T ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
100 |
0.25 |
ADME/T ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
600 |
0.22 |
ADME/T ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 2), Other |
Other |
60 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
480 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80178-2-O |
J774.A1 (Macrophage Cells) (cluster #2 Of 2), Other |
Other |
20 |
0.28 |
ADME/T ≤ 10μM |
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 3), Other |
Other |
60 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80193-3-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #3 Of 4), Other |
Other |
29 |
0.27 |
ADME/T ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
33 |
0.27 |
ADME/T ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
70 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80253-2-O |
MEF (Embryonic Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
3400 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
300 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2000 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80548-2-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
8 |
0.29 |
ADME/T ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 3), Other |
Other |
7400 |
0.18 |
ADME/T ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
900 |
0.22 |
ADME/T ≤ 10μM
|
|
Z80704-1-O |
BJ (Foreskin Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
2700 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80936-4-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #4 Of 4), Other |
Other |
64 |
0.26 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
420 |
0.23 |
ADME/T ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
ADME/T ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
7 |
0.29 |
ADME/T ≤ 10μM
|
|
Z81057-5-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #5 Of 5), Other |
Other |
30 |
0.27 |
ADME/T ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
400 |
0.23 |
ADME/T ≤ 10μM
|
|
Z81331-2-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
178 |
0.24 |
ADME/T ≤ 10μM
|
|
Q7ZJM1-1-V |
Human Immunodeficiency Virus Type 1 Integrase (cluster #1 Of 6), Viral |
Viruses |
900 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
-0.88 |
-58.15 |
8 |
12 |
1 |
208 |
544.533 |
5 |
↓
|
|
|
|
Analogs
-
3830631
-
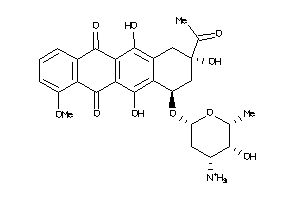
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-1-E |
Serine/threonine-protein Kinase Aurora-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.27 |
Binding ≤ 10μM |
|
MMP2-2-E |
72 KDa Type IV Collagenase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.21 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.22 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 2), Other |
Other |
2600 |
0.20 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
130 |
0.25 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
370 |
0.23 |
Functional ≤ 10μM
|
|
Z103219-1-O |
Ca9-22 (cluster #1 Of 1), Other |
Other |
1300 |
0.21 |
Functional ≤ 10μM
|
|
Z50347-3-O |
Saccharomyces Cerevisiae (cluster #3 Of 3), Other |
Other |
10000 |
0.18 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z80008-1-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80017-1-O |
A2780cisR (Cisplatin-resistant Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
6 |
0.30 |
Functional ≤ 10μM
|
|
Z80018-2-O |
A-375 (Malignant Melanoma Cells) (cluster #2 Of 4), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM |
|
Z80021-3-O |
A498 (Renal Carcinoma Cells) (cluster #3 Of 5), Other |
Other |
53 |
0.26 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
700 |
0.22 |
Functional ≤ 10μM |
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
310 |
0.23 |
Functional ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
8 |
0.29 |
Functional ≤ 10μM
|
|
Z80039-1-O |
BGC-823 (Stomach Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
1000 |
0.22 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80068-1-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
6000 |
0.19 |
Functional ≤ 10μM
|
|
Z80092-1-O |
CHO-K1 (Ovarian Cells) (cluster #1 Of 2), Other |
Other |
400 |
0.23 |
Functional ≤ 10μM
|
|
Z80105-2-O |
COR-L23 (Lung Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
319 |
0.23 |
Functional ≤ 10μM
|
|
Z80114-1-O |
Daudi (Burkitts Lymphoma Cells) (cluster #1 Of 4), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
6240 |
0.19 |
Functional ≤ 10μM |
|
Z80126-2-O |
DuPro (cluster #2 Of 2), Other |
Other |
1800 |
0.21 |
Functional ≤ 10μM
|
|
Z80138-1-O |
G-361 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
870 |
0.22 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
920 |
0.22 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
9320 |
0.18 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
92 |
0.25 |
Functional ≤ 10μM
|
|
Z80175-1-O |
IMR-32 (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
21 |
0.28 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
900 |
0.22 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
9700 |
0.18 |
Functional ≤ 10μM
|
|
Z80214-2-O |
M14 (Melanoma Cells) (cluster #2 Of 3), Other |
Other |
820 |
0.22 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
920 |
0.22 |
Functional ≤ 10μM
|
|
Z80227-1-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM
|
|
Z80240-1-O |
MDA-MB-361 (cluster #1 Of 1), Other |
Other |
33 |
0.27 |
Functional ≤ 10μM
|
|
Z80243-1-O |
MDA-MB-453 (Breast Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
120 |
0.25 |
Functional ≤ 10μM
|
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
7 |
0.29 |
Functional ≤ 10μM
|
|
Z80254-1-O |
MES-SA (Uterine Sarcoma Cells) (cluster #1 Of 2), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
704 |
0.22 |
Functional ≤ 10μM
|
|
Z80267-1-O |
MKN-1 (cluster #1 Of 1), Other |
Other |
640 |
0.22 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
3130 |
0.20 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z80295-6-O |
MT4 (Lymphocytes) (cluster #6 Of 8), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM |
|
Z80322-2-O |
NCI-H358 (Lung Carcinama Cells) (cluster #2 Of 2), Other |
Other |
620 |
0.22 |
Functional ≤ 10μM
|
|
Z80330-1-O |
NCI-N87 (cluster #1 Of 1), Other |
Other |
420 |
0.23 |
Functional ≤ 10μM
|
|
Z80354-4-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
35 |
0.27 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
830 |
0.22 |
Functional ≤ 10μM |
|
Z80364-1-O |
P388/ADR (Lymphoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80367-1-O |
P388/S (cluster #1 Of 1), Other |
Other |
13 |
0.28 |
Functional ≤ 10μM
|
|
Z80389-1-O |
PC-14 (cluster #1 Of 1), Other |
Other |
290 |
0.23 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
912 |
0.22 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
560 |
0.22 |
Functional ≤ 10μM
|
|
Z80441-1-O |
RXF 944 (cluster #1 Of 1), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
600 |
0.22 |
Functional ≤ 10μM |
|
Z80468-1-O |
SF-295 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
35 |
0.27 |
Functional ≤ 10μM
|
|
Z80472-1-O |
SiHa (Cervical Squamous Cell Carcinoma) (cluster #1 Of 2), Other |
Other |
500 |
0.23 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80477-1-O |
SK_HEP1 (Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
|
Z80479-1-O |
SK-MEL (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
1730 |
0.21 |
Functional ≤ 10μM
|
|
Z80481-1-O |
SK-MEL-1 (Metastatic Melanoma Cells) (cluster #1 Of 1), Other |
Other |
3092 |
0.20 |
Functional ≤ 10μM |
|
Z80482-3-O |
SK-MEL-2 (Melanoma Cells) (cluster #3 Of 4), Other |
Other |
97 |
0.25 |
Functional ≤ 10μM
|
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
600 |
0.22 |
Functional ≤ 10μM |
|
Z80490-3-O |
SK-MES-1 (cluster #3 Of 4), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80491-2-O |
SK-N-MC (Neuroepithelioma Cells) (cluster #2 Of 4), Other |
Other |
120 |
0.25 |
Functional ≤ 10μM
|
|
Z80492-1-O |
SK-N-SH (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
1100 |
0.21 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
800 |
0.22 |
Functional ≤ 10μM
|
|
Z80495-1-O |
SK-VLB (cluster #1 Of 1), Other |
Other |
6540 |
0.19 |
Functional ≤ 10μM
|
|
Z80503-1-O |
SNU1 (cluster #1 Of 2), Other |
Other |
4000 |
0.19 |
Functional ≤ 10μM
|
|
Z80509-1-O |
SNU-638 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
690 |
0.22 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
640 |
0.22 |
Functional ≤ 10μM
|
|
Z80559-1-O |
U251 (cluster #1 Of 3), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80560-1-O |
U-251 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80574-2-O |
UMUC3 (cluster #2 Of 2), Other |
Other |
9 |
0.29 |
Functional ≤ 10μM
|
|
Z80581-1-O |
VA13 (cluster #1 Of 1), Other |
Other |
699 |
0.22 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
5000 |
0.19 |
Functional ≤ 10μM
|
|
Z80595-1-O |
WIL2-NS (Lymphoblastoid Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.24 |
Functional ≤ 10μM |
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
44 |
0.26 |
Functional ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
9 |
0.29 |
Functional ≤ 10μM
|
|
Z80661-1-O |
A704 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.20 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
970 |
0.22 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
1000 |
0.22 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
72 |
0.26 |
Functional ≤ 10μM
|
|
Z80726-1-O |
TRAMP-C1A (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
76 |
0.26 |
Functional ≤ 10μM
|
|
Z80730-1-O |
C2D Cell Line (cluster #1 Of 1), Other |
Other |
47 |
0.26 |
Functional ≤ 10μM
|
|
Z80731-1-O |
C2G Cell Line (cluster #1 Of 1), Other |
Other |
29 |
0.27 |
Functional ≤ 10μM
|
|
Z80732-1-O |
TRAMP-C2H (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80735-1-O |
C38 (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z80768-1-O |
CH1 (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
6 |
0.30 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
520 |
0.23 |
Functional ≤ 10μM
|
|
Z80801-2-O |
CRL-7065 Cell Line (cluster #2 Of 2), Other |
Other |
500 |
0.23 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
368 |
0.23 |
Functional ≤ 10μM |
|
Z80846-1-O |
F460pv8/eto Cell Line (cluster #1 Of 2), Other |
Other |
20 |
0.28 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
50 |
0.26 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
60 |
0.26 |
Functional ≤ 10μM
|
|
Z80887-1-O |
H2981 Cell Line (cluster #1 Of 1), Other |
Other |
700 |
0.22 |
Functional ≤ 10μM
|
|
Z80889-1-O |
NCI-H322M (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
27 |
0.27 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
900 |
0.22 |
Functional ≤ 10μM |
|
Z80943-1-O |
HEL 299 (cluster #1 Of 1), Other |
Other |
1700 |
0.21 |
Functional ≤ 10μM |
|
Z80945-1-O |
HEp-2 (Laryngeal Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
59 |
0.26 |
Functional ≤ 10μM
|
|
Z81005-1-O |
HTB-54 Cell Line (cluster #1 Of 1), Other |
Other |
1100 |
0.21 |
Functional ≤ 10μM
|
|
Z81006-1-O |
Human Breast 401NL Tumor Cell Line (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
620 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
954 |
0.22 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
660 |
0.22 |
Functional ≤ 10μM
|
|
Z81025-1-O |
Human Lung LXF 529L Tumor Cell Line (cluster #1 Of 1), Other |
Other |
30 |
0.27 |
Functional ≤ 10μM
|
|
Z81026-1-O |
Human Lung LXF 629L Tumor Cell Line (cluster #1 Of 1), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM
|
|
Z81028-1-O |
Human Melanoma Cell Line (cluster #1 Of 1), Other |
Other |
280 |
0.24 |
Functional ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
10 |
0.29 |
Functional ≤ 10μM
|
|
Z81043-1-O |
N592 (cluster #1 Of 2), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
90 |
0.25 |
Functional ≤ 10μM
|
|
Z81101-1-O |
KBM-3 Cell Line (cluster #1 Of 1), Other |
Other |
26 |
0.27 |
Functional ≤ 10μM
|
|
Z81102-1-O |
KBM-3/DOX Cell Line (cluster #1 Of 1), Other |
Other |
490 |
0.23 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
7010 |
0.19 |
Functional ≤ 10μM
|
|
Z81133-1-O |
L1210 (1565) (cluster #1 Of 1), Other |
Other |
315 |
0.23 |
Functional ≤ 10μM |
|
Z81134-1-O |
L2987 (Lung Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
300 |
0.23 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81164-1-O |
LL Cell Line (cluster #1 Of 2), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81167-1-O |
LLTC Cell Line (cluster #1 Of 1), Other |
Other |
22 |
0.27 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
78 |
0.26 |
Functional ≤ 10μM
|
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
42 |
0.26 |
Functional ≤ 10μM
|
|
Z81201-1-O |
HOP-62 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
4 |
0.30 |
Functional ≤ 10μM
|
|
Z81211-1-O |
LXFL529 Cell Line (cluster #1 Of 1), Other |
Other |
40 |
0.27 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
79 |
0.25 |
Functional ≤ 10μM |
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
960 |
0.22 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
730 |
0.22 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
70 |
0.26 |
Functional ≤ 10μM
|
|
Z81257-1-O |
MEL-AC Cell Line (cluster #1 Of 1), Other |
Other |
1200 |
0.21 |
Functional ≤ 10μM
|
|
Z81261-1-O |
UACC-375 (cluster #1 Of 1), Other |
Other |
112 |
0.25 |
Functional ≤ 10μM
|
|
Z81277-2-O |
NCI-N417 (cluster #2 Of 2), Other |
Other |
230 |
0.24 |
Functional ≤ 10μM
|
|
Z81280-1-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
8 |
0.29 |
Functional ≤ 10μM
|
|
Z81284-1-O |
NCI-H647 (cluster #1 Of 2), Other |
Other |
1 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
80 |
0.25 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
820 |
0.22 |
Functional ≤ 10μM
|
|
Z81341-1-O |
Tumor Cell Lines (cluster #1 Of 1), Other |
Other |
74 |
0.26 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 2), Other |
Other |
500 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
100 |
0.25 |
ADME/T ≤ 10μM
|
|
Z80005-1-O |
3T3 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
3700 |
0.19 |
ADME/T ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
100 |
0.25 |
ADME/T ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
600 |
0.22 |
ADME/T ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 2), Other |
Other |
60 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
480 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80178-2-O |
J774.A1 (Macrophage Cells) (cluster #2 Of 2), Other |
Other |
20 |
0.28 |
ADME/T ≤ 10μM |
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 3), Other |
Other |
60 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80193-3-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #3 Of 4), Other |
Other |
29 |
0.27 |
ADME/T ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
33 |
0.27 |
ADME/T ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
70 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80253-2-O |
MEF (Embryonic Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
3400 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
300 |
0.23 |
ADME/T ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2000 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80548-2-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
8 |
0.29 |
ADME/T ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 3), Other |
Other |
7400 |
0.18 |
ADME/T ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
900 |
0.22 |
ADME/T ≤ 10μM
|
|
Z80704-1-O |
BJ (Foreskin Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
2700 |
0.20 |
ADME/T ≤ 10μM
|
|
Z80936-4-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #4 Of 4), Other |
Other |
64 |
0.26 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
420 |
0.23 |
ADME/T ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 1), Other |
Other |
10 |
0.29 |
ADME/T ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
7 |
0.29 |
ADME/T ≤ 10μM
|
|
Z81057-5-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #5 Of 5), Other |
Other |
30 |
0.27 |
ADME/T ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
400 |
0.23 |
ADME/T ≤ 10μM
|
|
Z81331-2-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
178 |
0.24 |
ADME/T ≤ 10μM
|
|
Q7ZJM1-1-V |
Human Immunodeficiency Virus Type 1 Integrase (cluster #1 Of 6), Viral |
Viruses |
900 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
-0.9 |
-63.19 |
8 |
12 |
1 |
208 |
544.533 |
5 |
↓
|
|
|
|
Analogs
-
31911781
-
-
3953819
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 48 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
Z104295-2-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #2 Of 2), Other |
Other |
590 |
0.31 |
Binding ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
2100 |
0.28 |
Functional ≤ 10μM
|
|
Z80021-4-O |
A498 (Renal Carcinoma Cells) (cluster #4 Of 5), Other |
Other |
57 |
0.36 |
Functional ≤ 10μM
|
|
Z80099-3-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
13 |
0.39 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
15 |
0.39 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
14 |
0.39 |
Functional ≤ 10μM
|
|
Z80712-4-O |
T47D (Breast Carcinoma Cells) (cluster #4 Of 7), Other |
Other |
16 |
0.39 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
34 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
13.34 |
-19.69 |
0 |
3 |
1 |
9 |
372.536 |
4 |
↓
|
|