|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.26 |
-1.89 |
-50.04 |
2 |
5 |
-1 |
93 |
154.101 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.20 |
1.08 |
-78.67 |
2 |
5 |
-1 |
93 |
154.101 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.89 |
1.56 |
-45.79 |
1 |
3 |
-1 |
56 |
160.152 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXDA-1-E |
D-amino-acid Oxidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
745 |
0.71 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
4.99 |
-46.52 |
1 |
3 |
-1 |
56 |
160.152 |
1 |
↓
|
|
|
|
Analogs
-
12410372
-
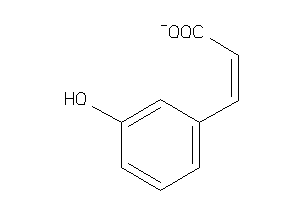
-
39258110
-
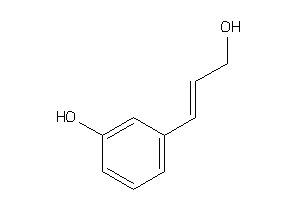
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50594-8-O |
Mus Musculus (cluster #8 Of 9), Other |
Other |
180 |
0.79 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
2.49 |
-50.04 |
1 |
3 |
-1 |
60 |
163.152 |
2 |
↓
|
|
|
|
Analogs
-
21803838
-
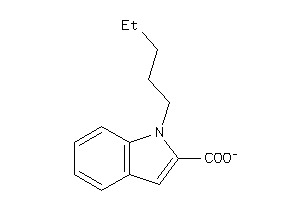
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
8100 |
0.55 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
8100 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
1.01 |
-48.72 |
0 |
3 |
-1 |
45 |
174.179 |
1 |
↓
|
|
|
|
Analogs
-
2005305
-
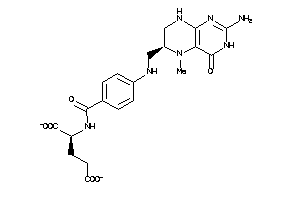
-
13509110
-
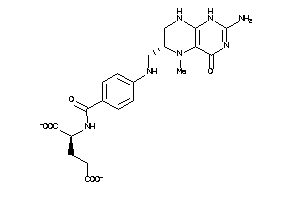
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.42 |
4.74 |
-137.95 |
6 |
13 |
-2 |
208 |
457.447 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
-2.42 |
5.55 |
-132.1 |
7 |
13 |
-1 |
210 |
458.455 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.42 |
4.87 |
-141.93 |
7 |
13 |
-1 |
210 |
458.455 |
9 |
↓
|
|
|
|
Analogs
-
4228235
-
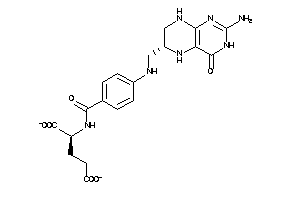
-
4228237
-
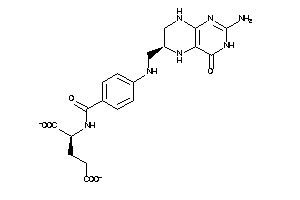
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.00 |
0.98 |
-145.01 |
7 |
13 |
-2 |
217 |
443.42 |
9 |
↓
|
|
|
|
Analogs
-
4228235
-

-
4228237
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.00 |
0.98 |
-145.17 |
7 |
13 |
-2 |
217 |
443.42 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.10 |
-1.46 |
-36.58 |
1 |
7 |
-1 |
96 |
195.158 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
-0.39 |
-1.31 |
-8.94 |
0 |
7 |
0 |
87 |
194.15 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.36 |
0.37 |
-10.78 |
2 |
7 |
0 |
93 |
196.166 |
0 |
↓
|
|
|
|
Analogs
-
22061777
-
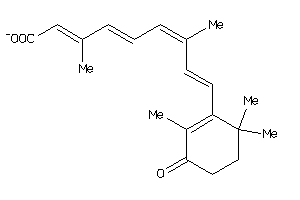
-
13415989
-
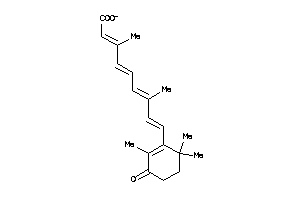
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.47 |
11.26 |
-62.61 |
0 |
3 |
-1 |
57 |
313.417 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.47 |
10.16 |
-14.64 |
1 |
3 |
0 |
54 |
314.425 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
8.49 |
-53.25 |
1 |
3 |
-1 |
60 |
315.433 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.56 |
7.41 |
-9.72 |
2 |
3 |
0 |
58 |
316.441 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.97 |
-3.36 |
-51.87 |
3 |
8 |
-1 |
136 |
475.558 |
8 |
↓
|
|
|
|
|
|
|
Analogs
-
6091356
-
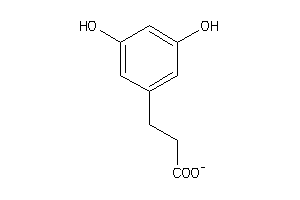
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.38 |
2.87 |
-47.31 |
1 |
3 |
-1 |
60 |
165.168 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
4.44 |
-47.65 |
1 |
3 |
-1 |
60 |
193.222 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
3.27 |
-47.74 |
2 |
4 |
-1 |
81 |
209.221 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.69 |
1.29 |
-10.33 |
3 |
4 |
0 |
78 |
210.229 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.76 |
-0.57 |
-101.57 |
3 |
13 |
-2 |
216 |
508.413 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.65 |
0.32 |
-50.75 |
4 |
10 |
-1 |
170 |
429.357 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.15 |
1.77 |
-52.94 |
1 |
7 |
-1 |
117 |
333.297 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q3I4V7-1-F |
Carbonic Anhydrase 2 (cluster #1 Of 4), Fungal |
Fungi |
900 |
0.77 |
Binding ≤ 10μM
|
|
Q5AJ71-2-F |
Carbonic Anhydrase (cluster #2 Of 4), Fungal |
Fungi |
1020 |
0.76 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
2.07 |
-47.22 |
1 |
3 |
-1 |
60 |
151.141 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.86 |
2.07 |
-48.56 |
1 |
3 |
-1 |
60 |
151.141 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
5012 |
0.62 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
5011.87234 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.39 |
0.9 |
-47.91 |
2 |
4 |
-1 |
81 |
167.14 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.32 |
4.83 |
-93.96 |
9 |
9 |
2 |
152 |
474.491 |
5 |
↓
|
|
|
|
Analogs
-
32149018
-
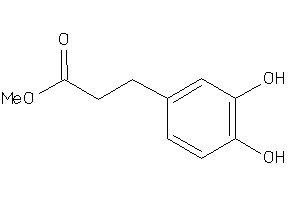
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.91 |
0.8 |
-48.42 |
2 |
4 |
-1 |
81 |
181.167 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
5.72 |
-46.16 |
0 |
2 |
-1 |
40 |
149.169 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
2.87 |
-46.42 |
1 |
3 |
-1 |
60 |
165.168 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYRO-7-F |
Tyrosinase (cluster #7 Of 8), Fungal |
Fungi |
1850 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
0.08 |
-48.08 |
2 |
4 |
-1 |
81 |
181.167 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.85 |
3.14 |
-47.38 |
1 |
4 |
-1 |
70 |
195.194 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
11.72 |
-60 |
1 |
4 |
-1 |
77 |
469.686 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.62 |
9.76 |
-14.05 |
2 |
4 |
0 |
75 |
470.694 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.62 |
9.8 |
-14.1 |
2 |
4 |
0 |
75 |
470.694 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
11.56 |
-61.01 |
1 |
4 |
-1 |
77 |
469.686 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.62 |
9.6 |
-12.63 |
2 |
4 |
0 |
75 |
470.694 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.62 |
9.59 |
-13.15 |
2 |
4 |
0 |
75 |
470.694 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
11.67 |
-60.91 |
1 |
4 |
-1 |
77 |
469.686 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.62 |
9.7 |
-12.43 |
2 |
4 |
0 |
75 |
470.694 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
11.21 |
-64.29 |
1 |
4 |
-1 |
77 |
469.686 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.62 |
9.26 |
-15.93 |
2 |
4 |
0 |
75 |
470.694 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.34 |
Binding ≤ 10μM
|
|
DHI2-1-E |
11-beta-hydroxysteroid Dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
257 |
0.27 |
Binding ≤ 10μM
|
|
KPCL-1-E |
Protein Kinase C Eta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
250 |
0.27 |
Binding ≤ 10μM |
|
PTN11-2-E |
Protein-tyrosine Phosphatase 2C (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9600 |
0.21 |
Binding ≤ 10μM
|
|
Z80418-3-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #3 Of 9), Other |
Other |
1300 |
0.24 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1_MOUSE |
P50172
|
11-beta-hydroxysteroid Dehydrogenase 1, Mouse |
10.58 |
0.33 |
Binding ≤ 1μM
|
|
DHI1_HUMAN |
P28845
|
11-beta-hydroxysteroid Dehydrogenase 1, Human |
11.78 |
0.33 |
Binding ≤ 1μM
|
|
DHI1_RAT |
P16232
|
11-beta-hydroxysteroid Dehydrogenase 1, Rat |
90 |
0.29 |
Binding ≤ 1μM
|
|
DHI2_HUMAN |
P80365
|
11-beta-hydroxysteroid Dehydrogenase 2, Human |
257 |
0.27 |
Binding ≤ 1μM
|
|
DHI2_RAT |
P50233
|
11-beta-hydroxysteroid Dehydrogenase 2, Rat |
360 |
0.27 |
Binding ≤ 1μM
|
|
KPCL_HUMAN |
P24723
|
Protein Kinase C Eta, Human |
250 |
0.27 |
Binding ≤ 1μM |
|
DHI1_RAT |
P16232
|
11-beta-hydroxysteroid Dehydrogenase 1, Rat |
90 |
0.29 |
Binding ≤ 10μM
|
|
DHI1_MOUSE |
P50172
|
11-beta-hydroxysteroid Dehydrogenase 1, Mouse |
10.58 |
0.33 |
Binding ≤ 10μM
|
|
DHI1_HUMAN |
P28845
|
11-beta-hydroxysteroid Dehydrogenase 1, Human |
11.78 |
0.33 |
Binding ≤ 10μM
|
|
DHI2_HUMAN |
P80365
|
11-beta-hydroxysteroid Dehydrogenase 2, Human |
257 |
0.27 |
Binding ≤ 10μM
|
|
DHI2_RAT |
P50233
|
11-beta-hydroxysteroid Dehydrogenase 2, Rat |
360 |
0.27 |
Binding ≤ 10μM
|
|
KPCL_HUMAN |
P24723
|
Protein Kinase C Eta, Human |
250 |
0.27 |
Binding ≤ 10μM |
|
PTN11_HUMAN |
Q06124
|
Protein-tyrosine Phosphatase 2C, Human |
9600 |
0.21 |
Binding ≤ 10μM
|
|
Z80418 |
Z80418
|
RAW264.7 (Monocytic-macrophage Leukemia Cells) |
1300 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
11.48 |
-62.34 |
1 |
4 |
-1 |
77 |
469.686 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
5.62 |
11.49 |
-61.44 |
1 |
4 |
-1 |
77 |
469.686 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.62 |
9.52 |
-14.08 |
2 |
4 |
0 |
75 |
470.694 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
11.56 |
-62.42 |
1 |
4 |
-1 |
77 |
469.686 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.62 |
9.59 |
-14.46 |
2 |
4 |
0 |
75 |
470.694 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
13109235
-
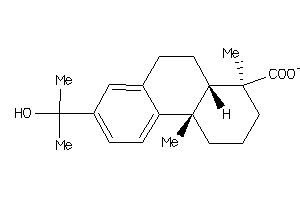
-
13109236
-
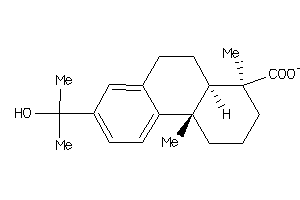
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
5.37 |
-51.49 |
2 |
4 |
-1 |
81 |
331.432 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.16 |
3.35 |
-10.8 |
3 |
4 |
0 |
78 |
332.44 |
2 |
↓
|
|
|
|
Analogs
-
13109235
-

-
13109236
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
5.9 |
-49.88 |
2 |
4 |
-1 |
81 |
331.432 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.16 |
3.85 |
-9.65 |
3 |
4 |
0 |
78 |
332.44 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.19 |
-5.83 |
-99.11 |
0 |
7 |
-2 |
115 |
272.234 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
1.63 |
-50.38 |
3 |
7 |
-1 |
135 |
299.214 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.47 |
2.41 |
-89.09 |
2 |
7 |
-2 |
138 |
298.206 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
5.75 |
-45.52 |
0 |
5 |
-1 |
68 |
239.247 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.43 |
-7.99 |
-44.83 |
1 |
5 |
-1 |
86 |
189.168 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.44 |
7.47 |
-51.52 |
1 |
4 |
-1 |
69 |
266.342 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.44 |
7.47 |
-51.79 |
1 |
4 |
-1 |
69 |
266.342 |
7 |
↓
|
|