|
|
Analogs
-
34485594
-
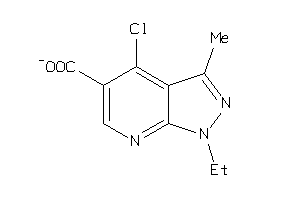
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
1.57 |
-7.08 |
0 |
5 |
0 |
57 |
253.689 |
3 |
↓
|
|
|
|
Analogs
-
32237759
-
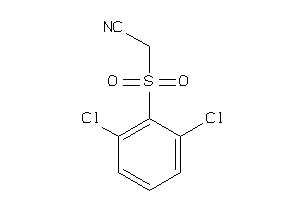
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
-1.5 |
-11.95 |
0 |
3 |
0 |
57 |
250.106 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.07 |
1.55 |
-6.13 |
1 |
5 |
0 |
59 |
184.195 |
4 |
↓
|
|
|
|
Analogs
-
39233921
-
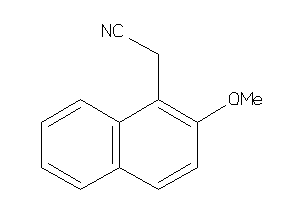
-
39281269
-
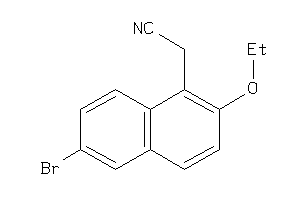
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
2.24 |
-8.41 |
0 |
2 |
0 |
33 |
276.133 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
8.07 |
-9.32 |
0 |
3 |
0 |
39 |
350.237 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.09 |
2.63 |
-9.96 |
1 |
4 |
0 |
59 |
164.164 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
1.35 |
-7.26 |
0 |
1 |
0 |
17 |
229.106 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
1.42 |
-7.9 |
0 |
1 |
0 |
17 |
229.106 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
-5.72 |
-12.04 |
3 |
4 |
0 |
60 |
271.345 |
2 |
↓
|
|
|
|
Analogs
-
34425901
-
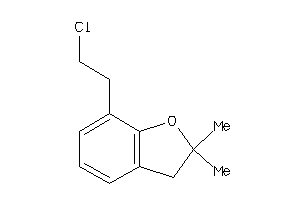
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
-1.19 |
-5.66 |
1 |
2 |
0 |
29 |
178.231 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
0.16 |
-9.68 |
0 |
5 |
0 |
59 |
310.419 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
2.36 |
-3.31 |
0 |
3 |
0 |
33 |
174.203 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
0.21 |
-7.9 |
0 |
3 |
0 |
30 |
202.044 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
0.39 |
-32.52 |
1 |
3 |
1 |
31 |
203.052 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
-6.12 |
-16.99 |
1 |
7 |
0 |
94 |
386.458 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.12 |
-5.56 |
-55.05 |
0 |
7 |
-1 |
96 |
385.45 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.12 |
-0.98 |
-45.07 |
4 |
5 |
1 |
63 |
173.24 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.12 |
-3.36 |
-10.41 |
3 |
5 |
0 |
62 |
172.232 |
1 |
↓
|
|
|
|
Analogs
-
42784466
-
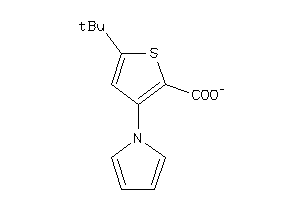
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
0.88 |
-52.48 |
0 |
3 |
-1 |
45 |
192.219 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
4.76 |
-48.57 |
0 |
3 |
-1 |
45 |
180.208 |
1 |
↓
|
|
|
|
Analogs
-
4426819
-
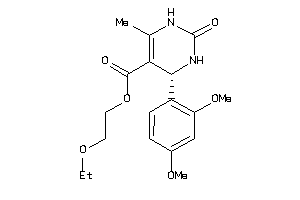
-
4426820
-
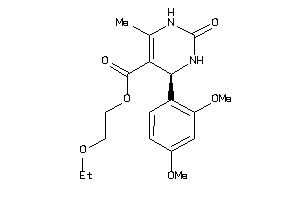
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
3.72 |
-11.73 |
2 |
8 |
0 |
95 |
350.371 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.46 |
1.78 |
-40.56 |
3 |
8 |
1 |
100 |
351.379 |
8 |
↓
|
|
|
|
Analogs
-
4426819
-

-
4426820
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
3.68 |
-13.1 |
2 |
8 |
0 |
95 |
350.371 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.46 |
1.24 |
-37.2 |
3 |
8 |
1 |
100 |
351.379 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
4.06 |
-14.12 |
3 |
4 |
0 |
74 |
238.703 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.70 |
4.29 |
-36.23 |
2 |
4 |
-1 |
72 |
237.695 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.70 |
4.24 |
-36.92 |
2 |
4 |
-1 |
72 |
237.695 |
3 |
↓
|
|
|
|
Analogs
-
34956906
-
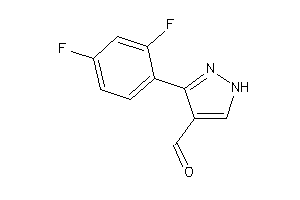
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
1.18 |
-10.17 |
1 |
2 |
0 |
28 |
208.211 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
7.26 |
-10.52 |
2 |
7 |
0 |
91 |
324.388 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
1.24 |
-15.27 |
2 |
5 |
0 |
75 |
268.338 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
0.84 |
-12.78 |
2 |
5 |
0 |
75 |
268.338 |
3 |
↓
|
|
|
|
Analogs
-
12367520
-
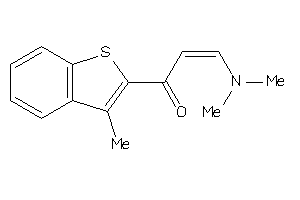
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
1.34 |
-8.75 |
0 |
2 |
0 |
20 |
245.347 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SERPH-1-E |
Collagen-binding Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.51 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SERPH_HUMAN |
P50454
|
Collagen-binding Protein 1, Human |
3100 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
3.99 |
-10.44 |
1 |
4 |
0 |
55 |
263.727 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SERPH-1-E |
Collagen-binding Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.51 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SERPH_HUMAN |
P50454
|
Collagen-binding Protein 1, Human |
3100 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
-2.15 |
-10.22 |
1 |
4 |
0 |
55 |
263.727 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
7.52 |
-4.45 |
1 |
3 |
0 |
36 |
294.398 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
7.47 |
-4.2 |
1 |
3 |
0 |
36 |
294.398 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.11 |
5.07 |
-9.03 |
0 |
3 |
0 |
33 |
213.283 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
8.48 |
-7.63 |
3 |
6 |
0 |
86 |
279.731 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
1.86 |
-5.98 |
2 |
3 |
0 |
44 |
231.295 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.04 |
4.32 |
-42.12 |
3 |
3 |
1 |
45 |
232.303 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
1.83 |
-6.1 |
2 |
3 |
0 |
44 |
231.295 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.04 |
4.32 |
-41.81 |
3 |
3 |
1 |
45 |
232.303 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
6.72 |
-3.19 |
0 |
1 |
0 |
3 |
199.297 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.80 |
9.18 |
-36.68 |
1 |
1 |
1 |
4 |
200.305 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
6.7 |
-3.23 |
0 |
1 |
0 |
3 |
199.297 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.80 |
9.17 |
-36.68 |
1 |
1 |
1 |
4 |
200.305 |
2 |
↓
|
|
|
|
Analogs
-
22064449
-
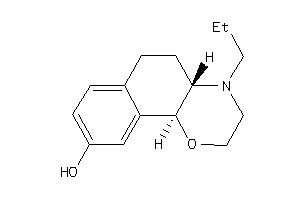
-
26824949
-
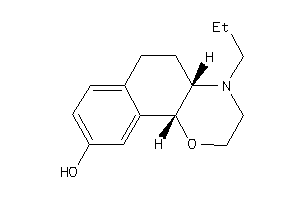
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
85 |
0.55 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
85 |
0.55 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
85 |
0.55 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
|
DRD4-3-E |
Dopamine D4 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.64 |
Functional ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
3 |
0.66 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
9 |
0.63 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
116 |
0.54 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
116 |
0.54 |
Binding ≤ 1μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
116 |
0.54 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
19.6 |
0.60 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
8.5 |
0.63 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
19.6 |
0.60 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
19.6 |
0.60 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
0.16 |
0.76 |
Binding ≤ 1μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
19.6 |
0.60 |
Binding ≤ 1μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
19.6 |
0.60 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
116 |
0.54 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
116 |
0.54 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
116 |
0.54 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
19.6 |
0.60 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
8.5 |
0.63 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
19.6 |
0.60 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
0.16 |
0.76 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
19.6 |
0.60 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
19.6 |
0.60 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
19.6 |
0.60 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
6.24 |
0.64 |
Functional ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
0.199526231 |
0.75 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
3.08 |
-8.58 |
1 |
3 |
0 |
33 |
247.338 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aR,10bR)-rel-4-Propyl-2,3,4,4a,5,10b-hexahydrochromeno[4,3-b][1,4]oxazin-9-ol
(4aR,10bR)-rel-4-Propyl-2,3,4,4a…
Find On:
PubMed —
Wikipedia —
Google
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
123 |
0.54 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
1.1 |
0.70 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
169 |
0.53 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
1180 |
0.46 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
123 |
0.54 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
1.1 |
0.70 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
169 |
0.53 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
1670 |
0.45 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
5.7 |
0.64 |
Functional ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
42 |
0.57 |
Functional ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
0.36 |
0.73 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
1.22 |
-5.81 |
1 |
4 |
0 |
42 |
249.31 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
1.06 |
-8.85 |
1 |
4 |
0 |
42 |
249.31 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.34 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.35 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
66 |
0.32 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.35 |
Binding ≤ 10μM
|
|
5HT1A-2-E |
Serotonin 1a (5-HT1a) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.37 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
89 |
0.32 |
Binding ≤ 10μM
|
|
Z80094-1-O |
CHO-MG (Ovarian Cells) (cluster #1 Of 1), Other |
Other |
7 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
89 |
0.32 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
18.6208714 |
0.35 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
322 |
0.29 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
4.5708819 |
0.38 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
16 |
0.35 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
0.1 |
0.45 |
Binding ≤ 1μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
0.158489319 |
0.44 |
Binding ≤ 1μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
24 |
0.34 |
Binding ≤ 1μM
|
|
5HT1D_MOUSE |
Q61224
|
Serotonin 1d (5-HT1d) Receptor, Mouse |
0.34 |
0.43 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
89 |
0.32 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
18.6208714 |
0.35 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
322 |
0.29 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
4.5708819 |
0.38 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
16 |
0.35 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
0.1 |
0.45 |
Binding ≤ 10μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
0.158489319 |
0.44 |
Binding ≤ 10μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
24 |
0.34 |
Binding ≤ 10μM
|
|
5HT1D_MOUSE |
Q61224
|
Serotonin 1d (5-HT1d) Receptor, Mouse |
0.34 |
0.43 |
Binding ≤ 10μM
|
|
5HT7R_HUMAN |
P34969
|
Serotonin 7 (5-HT7) Receptor, Human |
10000 |
0.23 |
Binding ≤ 10μM
|
|
Z80094 |
Z80094
|
CHO-MG (Ovarian Cells) |
7.1 |
0.37 |
Functional ≤ 10μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
7.1 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
11.38 |
-13.41 |
0 |
6 |
0 |
49 |
422.573 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
9.23 |
-13.26 |
1 |
6 |
0 |
60 |
408.546 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
9.03 |
-12.53 |
1 |
5 |
0 |
45 |
407.489 |
6 |
↓
|
|
|
|
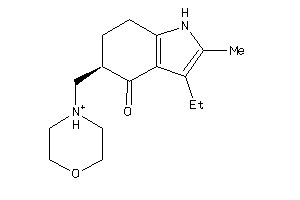
Analogs
-
512
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
265 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
5.69 |
-11.34 |
1 |
4 |
0 |
45 |
276.38 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.12 |
7.04 |
-48.98 |
1 |
4 |
1 |
43 |
277.388 |
3 |
↓
|
|
|
|
Analogs
-
512
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
265 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
5.69 |
-11.35 |
1 |
4 |
0 |
45 |
276.38 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.12 |
6.82 |
-50.27 |
1 |
4 |
1 |
43 |
277.388 |
3 |
↓
|
|
|
|
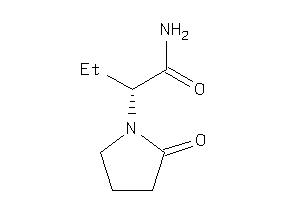
Analogs
-
3830990
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.23 |
1.97 |
-16.36 |
2 |
4 |
0 |
63 |
170.212 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
-0.02 |
0.04 |
-9.17 |
2 |
4 |
0 |
64 |
170.212 |
3 |
↓
|
|
|
|
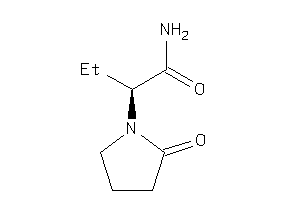
Analogs
-
1547851
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.23 |
2 |
-16.44 |
2 |
4 |
0 |
63 |
170.212 |
3 |
↓
|
|
|
|
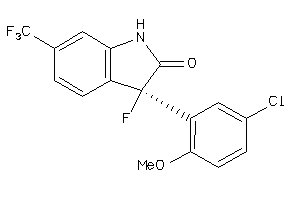
Analogs
-
538646
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
2.86 |
-8.57 |
1 |
3 |
0 |
38 |
359.706 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.43 |
Binding ≤ 10μM
|
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.02 |
2.73 |
-7.89 |
0 |
4 |
0 |
33 |
364.537 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
JUN-1-E |
Proto-oncogene C-JUN (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2030 |
0.35 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
6.95 |
-11.64 |
4 |
3 |
0 |
76 |
335.354 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.06 |
6.73 |
-38.57 |
5 |
3 |
1 |
77 |
336.362 |
4 |
↓
|
|