|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.99 |
-10.86 |
6 |
7 |
0 |
129 |
397.467 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.14 |
7.8 |
-55.6 |
7 |
7 |
1 |
131 |
398.475 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.14 |
7.83 |
-30.11 |
7 |
7 |
1 |
131 |
398.475 |
3 |
↓
|
|
|
|
Analogs
-
18275342
-
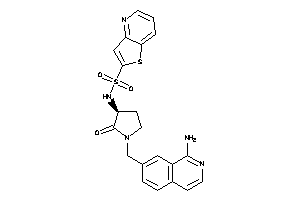
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
4.39 |
-24.78 |
3 |
8 |
0 |
118 |
453.549 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.92 |
5.12 |
-61.15 |
2 |
8 |
-1 |
120 |
452.541 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.92 |
4.85 |
-44.67 |
4 |
8 |
1 |
120 |
454.557 |
5 |
↓
|
|
|
|
Analogs
-
39048538
-
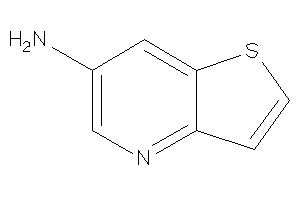
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
1.91 |
-6.18 |
2 |
2 |
0 |
39 |
150.206 |
0 |
↓
|
|
|
|
Analogs
-
28525510
-
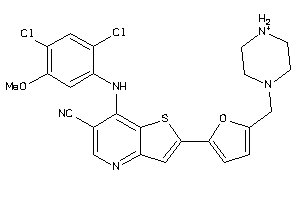
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
10.41 |
-44.41 |
2 |
7 |
1 |
79 |
529.473 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.92 |
10.54 |
-44.8 |
2 |
7 |
1 |
79 |
529.473 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.92 |
8.07 |
-9.91 |
1 |
7 |
0 |
78 |
528.465 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
56 |
0.29 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.08 |
9.29 |
-45.83 |
2 |
7 |
1 |
79 |
546.529 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.08 |
6.82 |
-10.57 |
1 |
7 |
0 |
77 |
545.521 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.08 |
9.15 |
-48.03 |
2 |
7 |
1 |
79 |
546.529 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
13 |
0.31 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Functional ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Functional ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.22 |
Functional ≤ 10μM
|
|
MP2K1-2-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.23 |
Functional ≤ 10μM
|
|
RAF1-2-E |
Serine/threonine-protein Kinase RAF (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.23 |
Functional ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.40 |
10.31 |
-44.6 |
2 |
7 |
1 |
79 |
540.5 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.40 |
10.19 |
-49.45 |
2 |
7 |
1 |
79 |
540.5 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.40 |
7.84 |
-10.43 |
1 |
7 |
0 |
77 |
539.492 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
51 |
0.28 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.47 |
10.33 |
-44.89 |
2 |
7 |
1 |
79 |
540.5 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.47 |
10.19 |
-42.68 |
2 |
7 |
1 |
79 |
540.5 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.47 |
7.86 |
-11.29 |
1 |
7 |
0 |
77 |
539.492 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.29 |
9.86 |
-44.91 |
2 |
6 |
1 |
66 |
487.436 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.29 |
7.39 |
-9.35 |
1 |
6 |
0 |
64 |
486.428 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
47 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.42 |
11.33 |
-43.26 |
2 |
6 |
1 |
69 |
510.474 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.42 |
11.21 |
-48.43 |
2 |
6 |
1 |
69 |
510.474 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.42 |
8.86 |
-8.15 |
1 |
6 |
0 |
68 |
509.466 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
6.33 |
-17.92 |
3 |
6 |
0 |
79 |
411.434 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.92 |
6.73 |
-42.56 |
4 |
6 |
1 |
80 |
412.442 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
6.67 |
-18.55 |
3 |
6 |
0 |
79 |
411.434 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.84 |
7.32 |
-38.99 |
4 |
6 |
1 |
80 |
412.442 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.52 |
9.83 |
-10.59 |
1 |
3 |
0 |
42 |
404.438 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.57 |
9.13 |
-11.97 |
1 |
4 |
0 |
51 |
434.464 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.27 |
10.25 |
-13.12 |
1 |
4 |
0 |
66 |
429.448 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
9.97 |
-9.53 |
1 |
3 |
0 |
42 |
418.465 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.22 |
10.23 |
-33.33 |
2 |
3 |
1 |
43 |
419.473 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
-0.68 |
-10.85 |
0 |
4 |
0 |
42 |
398.431 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.91 |
-0.58 |
-36.81 |
1 |
4 |
1 |
43 |
399.439 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
3.28 |
-8.61 |
1 |
3 |
0 |
42 |
260.24 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
3.46 |
-57.09 |
0 |
4 |
-1 |
62 |
208.218 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.78 |
2.54 |
-11.25 |
1 |
4 |
0 |
59 |
209.226 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
4.5 |
-9.31 |
3 |
6 |
0 |
86 |
329.381 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACACA-1-E |
Acetyl-CoA Carboxylase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.29 |
Binding ≤ 10μM
|
|
ACACB-1-E |
Acetyl-CoA Carboxylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.29 |
Binding ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
49 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
5.67 |
-22.68 |
2 |
9 |
0 |
98 |
554.639 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.62 |
8.04 |
-66.35 |
3 |
9 |
1 |
99 |
555.647 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
5.96 |
-7.03 |
3 |
5 |
0 |
77 |
396.241 |
4 |
↓
|
|
|
|
Analogs
-
19834575
-
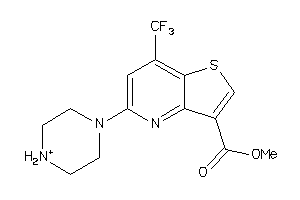
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.75 |
13.53 |
-44.64 |
1 |
5 |
1 |
47 |
450.506 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.75 |
11.3 |
-11.14 |
0 |
5 |
0 |
46 |
449.498 |
6 |
↓
|
|
|
|
Analogs
-
19834575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.84 |
14.78 |
-44.64 |
1 |
5 |
1 |
47 |
478.56 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.84 |
12.56 |
-10.86 |
0 |
5 |
0 |
46 |
477.552 |
7 |
↓
|
|
|
|
Analogs
-
19834575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.24 |
14.29 |
-44.61 |
1 |
5 |
1 |
47 |
464.533 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.24 |
12.08 |
-11.01 |
0 |
5 |
0 |
46 |
463.525 |
7 |
↓
|
|
|
|
Analogs
-
19834575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
13.28 |
-49.29 |
1 |
6 |
1 |
56 |
480.532 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.88 |
11.08 |
-13.09 |
0 |
6 |
0 |
55 |
479.524 |
9 |
↓
|
|
|
|
Analogs
-
19834575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
8.5 |
-19.51 |
1 |
9 |
0 |
93 |
496.515 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.89 |
10.64 |
-56.82 |
2 |
9 |
1 |
94 |
497.523 |
7 |
↓
|
|
|
|
Analogs
-
19834575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
9.18 |
-11.85 |
0 |
5 |
0 |
46 |
397.422 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.90 |
11.43 |
-42.66 |
1 |
5 |
1 |
47 |
398.43 |
5 |
↓
|
|
|
|
Analogs
-
19834587
-
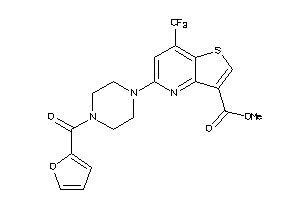
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
12.86 |
-53.1 |
1 |
8 |
1 |
86 |
498.503 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.03 |
10.62 |
-14.43 |
0 |
8 |
0 |
85 |
497.495 |
9 |
↓
|
|
|
|
Analogs
-
19834575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
8.67 |
-16.18 |
1 |
8 |
0 |
99 |
509.535 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.56 |
10.82 |
-54.44 |
2 |
8 |
1 |
100 |
510.543 |
7 |
↓
|
|
|
|
Analogs
-
19834575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
11.02 |
-14.45 |
0 |
6 |
0 |
69 |
460.481 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.08 |
13.24 |
-57.16 |
1 |
6 |
1 |
71 |
461.489 |
6 |
↓
|
|
|
|
Analogs
-
19834575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
8.83 |
-12.8 |
0 |
7 |
0 |
72 |
431.436 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.16 |
10.97 |
-45.08 |
1 |
7 |
1 |
73 |
432.444 |
8 |
↓
|
|
|
|
Analogs
-
19834575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.25 |
14.48 |
-47.52 |
1 |
5 |
1 |
47 |
464.533 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.25 |
12.27 |
-11.91 |
0 |
5 |
0 |
46 |
463.525 |
8 |
↓
|
|
|
|
Analogs
-
19834575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.28 |
14.26 |
-48.12 |
1 |
5 |
1 |
47 |
462.517 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.28 |
12.02 |
-12.34 |
0 |
5 |
0 |
46 |
461.509 |
7 |
↓
|
|
|
|
Analogs
-
19834587
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
11.93 |
-53.34 |
1 |
8 |
1 |
86 |
484.476 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.65 |
9.7 |
-14.63 |
0 |
8 |
0 |
85 |
483.468 |
8 |
↓
|
|