|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
8.89 |
-15.45 |
2 |
5 |
0 |
75 |
402.56 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.90 |
11.55 |
-15.36 |
1 |
5 |
0 |
66 |
430.614 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
9.25 |
-9.56 |
1 |
3 |
0 |
53 |
282.318 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.38 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
520 |
0.38 |
Binding ≤ 1μM
|
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
520 |
0.38 |
Binding ≤ 10μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
520 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
7.57 |
-22.01 |
2 |
5 |
0 |
70 |
312.369 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.16 |
8.79 |
-64.49 |
1 |
5 |
-1 |
72 |
311.361 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
25 |
0.44 |
Binding ≤ 1μM
|
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
25 |
0.44 |
Binding ≤ 10μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
25 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
6.76 |
-17.75 |
3 |
5 |
0 |
78 |
326.396 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.47 |
7.98 |
-62.49 |
2 |
5 |
-1 |
81 |
325.388 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.17 |
14.44 |
-33.76 |
2 |
4 |
1 |
46 |
396.53 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
15.55 |
-44.54 |
2 |
4 |
1 |
46 |
499.517 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
16.02 |
-33.91 |
2 |
4 |
1 |
46 |
499.517 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
4.17 |
-21.23 |
4 |
6 |
0 |
93 |
327.384 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
7.6 |
-16.89 |
3 |
6 |
0 |
79 |
369.465 |
8 |
↓
|
|
|
|
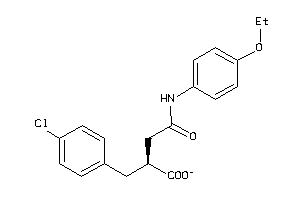
Analogs
-
5199501
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
10.29 |
-54.79 |
1 |
5 |
-1 |
78 |
360.817 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.92 |
8.5 |
-16 |
2 |
5 |
0 |
76 |
361.825 |
8 |
↓
|
|
|
|
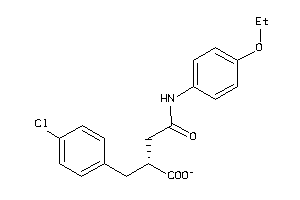
Analogs
-
5199497
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
10.25 |
-58.33 |
1 |
5 |
-1 |
78 |
360.817 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.92 |
8.51 |
-15.84 |
2 |
5 |
0 |
76 |
361.825 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
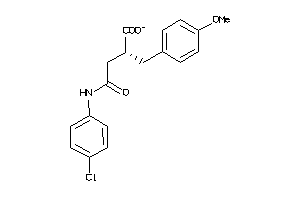
Analogs
-
5199522
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
0.56 |
-58.59 |
1 |
5 |
-1 |
78 |
346.79 |
7 |
↓
|
|
|
|
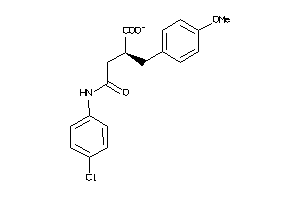
Analogs
-
5199518
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
0.55 |
-56.93 |
1 |
5 |
-1 |
78 |
346.79 |
7 |
↓
|
|
|
|
Analogs
-
351295
-
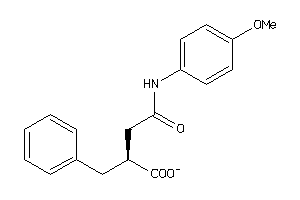
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
1.33 |
-61.87 |
1 |
5 |
-1 |
78 |
340.399 |
7 |
↓
|
|
|
|
Analogs
-
351295
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
1.44 |
-59.6 |
1 |
5 |
-1 |
78 |
340.399 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
7.28 |
-19.22 |
1 |
4 |
0 |
63 |
386.3 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
9.35 |
-17.62 |
2 |
4 |
0 |
58 |
358.869 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
8.72 |
-10.56 |
1 |
4 |
0 |
48 |
378.266 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.27 |
9.33 |
-15.77 |
1 |
5 |
0 |
59 |
386.517 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.27 |
9.15 |
-17.54 |
1 |
5 |
0 |
59 |
386.517 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
5.67 |
-15.88 |
4 |
5 |
0 |
84 |
325.412 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
5 |
-15.81 |
4 |
5 |
0 |
84 |
311.385 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
6.07 |
-11.48 |
3 |
4 |
0 |
64 |
284.359 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
11 |
-17.46 |
2 |
4 |
0 |
58 |
352.478 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
5.99 |
-14.61 |
2 |
3 |
0 |
49 |
273.307 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.58 |
6.75 |
-55.11 |
1 |
3 |
-1 |
52 |
272.299 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.64 |
3.34 |
-62.8 |
5 |
4 |
1 |
77 |
289.33 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.64 |
3.04 |
-13.01 |
4 |
4 |
0 |
75 |
288.322 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.64 |
3.34 |
-63.46 |
5 |
4 |
1 |
77 |
289.33 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.64 |
3 |
-14.94 |
4 |
4 |
0 |
75 |
288.322 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
6.11 |
-12.98 |
2 |
3 |
0 |
49 |
269.344 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
6.85 |
-11 |
2 |
3 |
0 |
49 |
269.344 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
6.02 |
-9.88 |
2 |
3 |
0 |
49 |
273.307 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
6.48 |
-12.21 |
2 |
3 |
0 |
49 |
289.762 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.09 |
7.24 |
-55.8 |
1 |
3 |
-1 |
52 |
288.754 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
6.55 |
-9.58 |
2 |
3 |
0 |
49 |
289.762 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
11.33 |
-10.71 |
0 |
2 |
0 |
20 |
267.372 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
6.4 |
-12.2 |
2 |
3 |
0 |
49 |
269.344 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.45 |
9.96 |
-13.75 |
1 |
2 |
0 |
29 |
263.34 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
7.6 |
-10.61 |
3 |
3 |
0 |
55 |
268.36 |
5 |
↓
|
|