|
|
Analogs
-
3789310
-
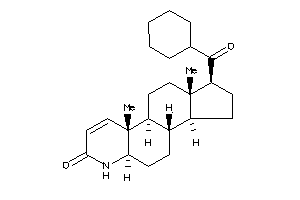
-
3789315
-
-
3791185
-
-
3791186
-
-
5440632
-
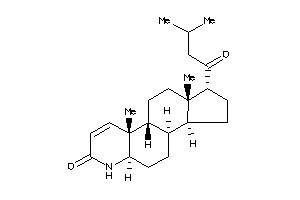
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bS,5aR,9aR,9bS,11aS)-1-butyryl-9a,11a-dimethyl-1,2,3,3a,3b,4,5,5a,6,9b,10,11-dodecahydroind
(1R,3aS,3bS,5aR,9aR,9bS,11aS)-1-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
0.4 |
-10.86 |
1 |
3 |
0 |
46 |
343.511 |
3 |
↓
|
|
|
|
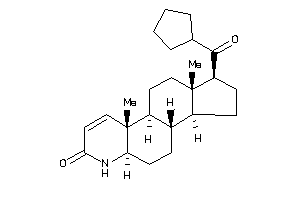
Analogs
-
40786689
-
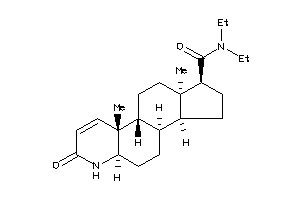
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
0.11 |
-13.69 |
1 |
4 |
0 |
49 |
372.553 |
3 |
↓
|
|
|
|
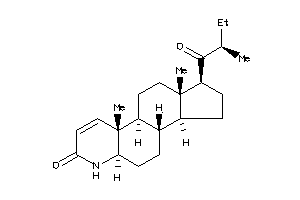
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-[(1R,3aS,3bR,5aR,9aR,9bR,11aR)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,9b,10,11-decahydro-1H-ind
2-[(1R,3aS,3bR,5aR,9aR,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
11.49 |
-16.04 |
2 |
6 |
0 |
78 |
488.676 |
4 |
↓
|
|
|
|
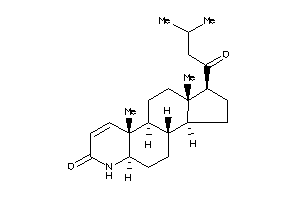
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-[(1R,3aS,3bR,5aR,9aR,9bR,11aR)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,9b,10,11-decahydro-1H-ind
2-[(1R,3aS,3bR,5aR,9aR,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
11.49 |
-16.13 |
2 |
6 |
0 |
78 |
488.676 |
4 |
↓
|
|
|
|
Analogs
-
36152975
-
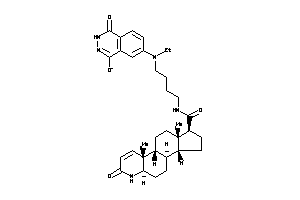
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-N-[4-[(1,4-dioxo-2,3-dihydrophthalazin-6-yl)-ethyl-amino]butyl]-9a,11a
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-N-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.21 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
6.72 |
-59.32 |
3 |
9 |
-1 |
130 |
574.746 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.58 |
5.99 |
-22.01 |
4 |
9 |
0 |
127 |
575.754 |
8 |
↓
|
|
|
|
Analogs
-
36152968
-
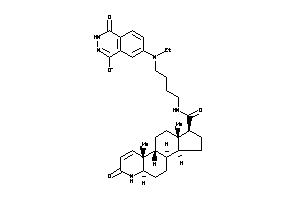
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-N-[4-[(1,4-dioxo-2,3-dihydrophthalazin-6-yl)-ethyl-amino]butyl]-9a,11a
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-N-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.21 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
6.3 |
-60.93 |
3 |
9 |
-1 |
130 |
574.746 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.58 |
5.57 |
-23.66 |
4 |
9 |
0 |
127 |
575.754 |
8 |
↓
|
|
|
|
Analogs
-
36153160
-
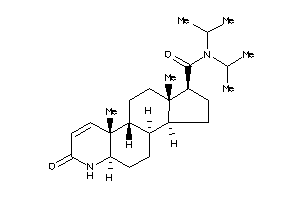
-
36153165
-
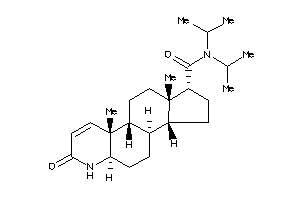
-
36153169
-
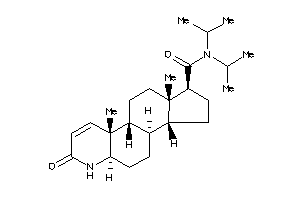
-
40786689
-

-
40786691
-
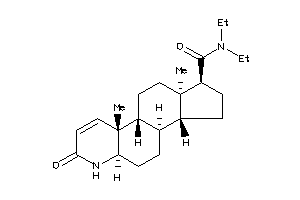
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-N,N-diisopropyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11-do
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
10.05 |
-13.13 |
1 |
4 |
0 |
49 |
400.607 |
3 |
↓
|
|
|
|
Analogs
-
36153165
-

-
36153169
-

-
40786689
-

-
40786691
-

-
45298407
-
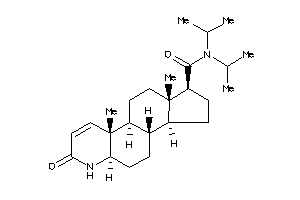
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-N,N-diisopropyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11-do
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
9.79 |
-11.98 |
1 |
4 |
0 |
49 |
400.607 |
3 |
↓
|
|
|
|
Analogs
-
36153169
-

-
40786689
-

-
40786691
-

-
45298407
-

-
36153156
-
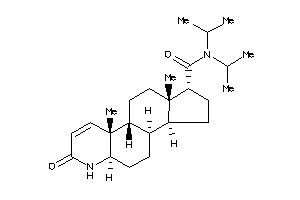
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-N,N-diisopropyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11-do
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
9.12 |
-8.32 |
1 |
4 |
0 |
49 |
400.607 |
3 |
↓
|
|
|
|
Analogs
-
40786689
-

-
40786691
-

-
45298407
-

-
36153156
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-N,N-diisopropyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11-do
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
9.6 |
-12.73 |
1 |
4 |
0 |
49 |
400.607 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bS,5aR,9aS,9bR,11aR)-1-hydroxy-6,9a,11a-trimethyl-2,3,3a,3b,4,5,5a,9b,10,11-decahydro-1H-in
(1R,3aS,3bS,5aR,9aS,9bR,11aR)-1-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
6.02 |
-9.56 |
1 |
3 |
0 |
41 |
303.446 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2R,3aS,3bS,5aR,9aS,9bR,11aR)-1-hydroxy-2,6,9a,11a-tetramethyl-2,3,3a,3b,4,5,5a,9b,10,11-decahydr
(1R,2R,3aS,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
31 |
0.46 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
119 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
6.8 |
-9.38 |
1 |
3 |
0 |
41 |
317.473 |
0 |
↓
|
|
|
|
Analogs
-
42964629
-
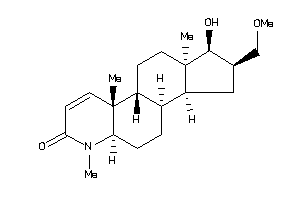
-
42964632
-
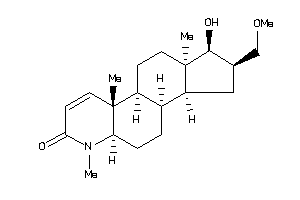
-
42964635
-
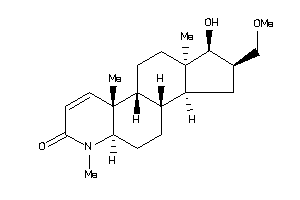
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2S,3aS,3bS,5aR,9aS,9bR,11aR)-1-hydroxy-2-(methoxymethyl)-6,9a,11a-trimethyl-2,3,3a,3b,4,5,5a,9b,
(1R,2S,3aS,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
24 |
0.43 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
612 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
5.8 |
-11.63 |
1 |
4 |
0 |
50 |
347.499 |
2 |
↓
|
|
|
|
Analogs
-
42990153
-
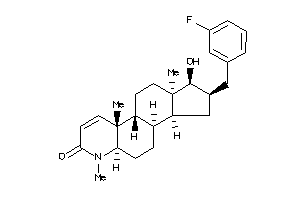
-
42990154
-
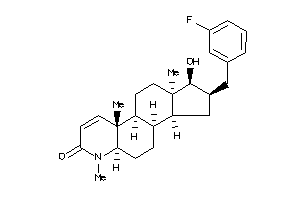
-
42990156
-
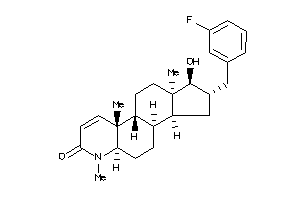
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2S,3aS,3bS,5aR,9aS,9bR,11aR)-2-[(3-fluorophenyl)methyl]-1-hydroxy-6,9a,11a-trimethyl-2,3,3a,3b,4
(1R,2S,3aS,3bS,5aR,9aS,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
23 |
0.36 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
240 |
0.31 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
14 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.98 |
10.33 |
-12.07 |
1 |
3 |
0 |
41 |
411.561 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
8.73 |
-11.04 |
1 |
4 |
0 |
55 |
359.51 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
8.31 |
-10.35 |
1 |
4 |
0 |
55 |
359.51 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
8.69 |
-12.05 |
1 |
4 |
0 |
55 |
359.51 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
8.83 |
-11.53 |
1 |
4 |
0 |
55 |
359.51 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-1-(3-methylbutanoyl)-2,3,3a,3b,4,5,5a,9b,10,11-deca
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
11.85 |
-10.93 |
0 |
3 |
0 |
37 |
371.565 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-1-(3-methylbutanoyl)-2,3,3a,3b,4,5,5a,9b,10,11-deca
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
11.82 |
-11.39 |
0 |
3 |
0 |
37 |
371.565 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-1-(3-methylbutanoyl)-2,3,3a,3b,4,5,5a,9b,10,11-deca
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
11.16 |
-9.46 |
0 |
3 |
0 |
37 |
371.565 |
3 |
↓
|
|
|
|
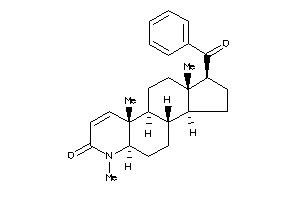
Analogs
-
6144177
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-1-(3-methylbutanoyl)-2,3,3a,3b,4,5,5a,9b,10,11-deca
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
11.57 |
-12.02 |
0 |
3 |
0 |
37 |
371.565 |
3 |
↓
|
|
|
|
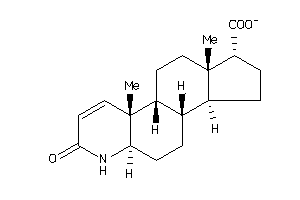
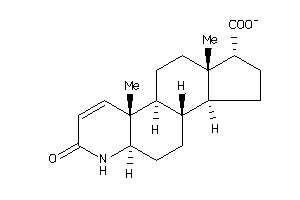
Analogs
-
22065515
-
-
22065518
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S)-2-[(1S,3aS,3bR,5aR,9aR,9bR,11aS)-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11-dodecahydr
(2S)-2-[(1S,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
8.49 |
-52.82 |
1 |
4 |
-1 |
69 |
344.475 |
2 |
↓
|
|
|
|
Analogs
-
22065515
-

-
22065518
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R)-2-[(1S,3aS,3bR,5aR,9aR,9bR,11aS)-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11-dodecahydr
(2R)-2-[(1S,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
8.24 |
-51.49 |
1 |
4 |
-1 |
69 |
344.475 |
2 |
↓
|
|
|
|
Analogs
-
22065515
-

-
22065518
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S)-2-[(1R,3aS,3bR,5aR,9aR,9bR,11aS)-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11-dodecahydr
(2S)-2-[(1R,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
8.66 |
-53.31 |
1 |
4 |
-1 |
69 |
344.475 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R)-2-[(1R,3aS,3bR,5aR,9aR,9bR,11aS)-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11-dodecahydr
(2R)-2-[(1R,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
8.7 |
-52.75 |
1 |
4 |
-1 |
69 |
344.475 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aR)-6,9a,11a-trimethyl-7-oxo-N-(1,1,3,3-tetramethylbutyl)-2,3,3a,3b,4,5,5a
(1S,3aS,3bR,5aR,9aR,9bR,11aR)-6,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.84 |
12.65 |
-11.2 |
1 |
4 |
0 |
49 |
442.688 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-N-(1,1,3,3-tetramethylbutyl)-2,3,3a,3b,4,5,5a
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.84 |
13.09 |
-10.31 |
1 |
4 |
0 |
49 |
442.688 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bS,11aR)-6,9a,11a-trimethyl-7-oxo-N-(1,1,3,3-tetramethylbutyl)-2,3,3a,3b,4,5,5a
(1S,3aS,3bR,5aR,9aR,9bS,11aR)-6,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.84 |
12.84 |
-12.25 |
1 |
4 |
0 |
49 |
442.688 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bS,11aS)-6,9a,11a-trimethyl-7-oxo-N-(1,1,3,3-tetramethylbutyl)-2,3,3a,3b,4,5,5a
(1S,3aS,3bR,5aR,9aR,9bS,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.84 |
12.96 |
-10.69 |
1 |
4 |
0 |
49 |
442.688 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aR)-N,N-diethyl-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,9b,10,11-decahyd
(1S,3aS,3bR,5aR,9aR,9bR,11aR)-N,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
10.89 |
-13.27 |
0 |
4 |
0 |
41 |
386.58 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bR,11aR)-N,N-diethyl-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,9b,10,11-decahyd
(1S,3aR,3bR,5aR,9aR,9bR,11aR)-N,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
10.79 |
-13.51 |
0 |
4 |
0 |
41 |
386.58 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S)-4-[(1R,3aS,3bS,4R,5aR,9aS,9bS,11aS)-4-ethyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11
(4S)-4-[(1R,3aS,3bS,4R,5aR,9aS,9…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
10.35 |
-51.98 |
1 |
4 |
-1 |
69 |
400.583 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.22 |
8.38 |
-10.94 |
2 |
4 |
0 |
66 |
401.591 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S)-4-[(1R,3aS,3bS,4S,5aR,9aS,9bS,11aS)-4-ethyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11
(4S)-4-[(1R,3aS,3bS,4S,5aR,9aS,9…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
11.37 |
-50.24 |
1 |
4 |
-1 |
69 |
400.583 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.22 |
9.39 |
-12.29 |
2 |
4 |
0 |
66 |
401.591 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S)-4-[(1R,3aS,3bS,4R,5aS,9aS,9bS,11aS)-4-ethyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11
(4S)-4-[(1R,3aS,3bS,4R,5aS,9aS,9…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
10.41 |
-52.59 |
1 |
4 |
-1 |
69 |
400.583 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.22 |
8.44 |
-10.79 |
2 |
4 |
0 |
66 |
401.591 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S)-4-[(1R,3aS,3bS,4S,5aS,9aS,9bS,11aS)-4-ethyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,9b,10,11
(4S)-4-[(1R,3aS,3bS,4S,5aS,9aS,9…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
11.26 |
-50.43 |
1 |
4 |
-1 |
69 |
400.583 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.22 |
9.28 |
-11.95 |
2 |
4 |
0 |
66 |
401.591 |
5 |
↓
|
|