|
|
Analogs
-
4095715
-
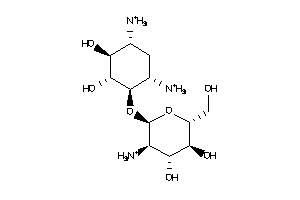
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TARB1-1-E |
TAR RNA Binding Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Binding ≤ 10μM
|
|
TRBP2-1-E |
TAR RNA Binding Protein 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Binding ≤ 10μM
|
|
Z100263-1-O |
Ribosomal RNA A-site (cluster #1 Of 1), Other |
Other |
7800 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.26 |
-14.91 |
-216.63 |
15 |
10 |
3 |
208 |
325.386 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.26 |
-15.78 |
-44.61 |
13 |
10 |
1 |
205 |
323.37 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.26 |
-15.37 |
-101.49 |
14 |
10 |
2 |
207 |
324.378 |
3 |
↓
|
|
|
|
Analogs
-
34319463
-
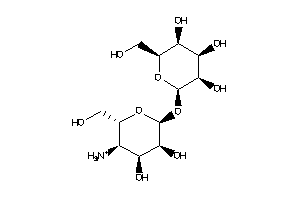
-
34319464
-
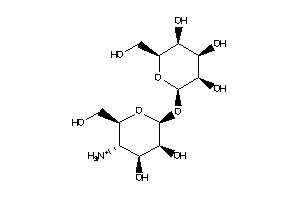
-
34319465
-
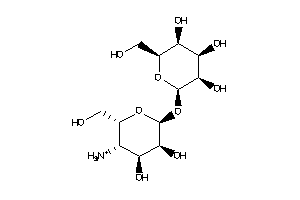
-
31545286
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2R,4S,5S)-6-[(2S,3R,4S,5S,6R)-3-amino-4,5,6-trihydroxy-tetrahydropyran-2-yl]oxycyclohexane-1,2,3
(1R,2R,4S,5S)-6-[(2S,3R,4S,5S,6R…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLCB3-1-E |
Phospholipase C-beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
PLCG2-1-E |
Phospholipase C-gamma-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.81 |
-15.04 |
-44.29 |
11 |
11 |
1 |
208 |
328.294 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.81 |
-16.32 |
-14.1 |
10 |
11 |
0 |
206 |
327.286 |
2 |
↓
|
|
|
|
Analogs
-
34319463
-

-
34319464
-

-
34319465
-

-
31545286
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2R,4S,5S)-6-[(2S,3S,4S,5S,6R)-3-amino-4,5,6-trihydroxy-tetrahydropyran-2-yl]oxycyclohexane-1,2,3
(1R,2R,4S,5S)-6-[(2S,3S,4S,5S,6R…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLCB3-1-E |
Phospholipase C-beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
PLCG2-1-E |
Phospholipase C-gamma-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.81 |
-16.79 |
-51.6 |
11 |
11 |
1 |
208 |
328.294 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.81 |
-17.19 |
-13.67 |
10 |
11 |
0 |
206 |
327.286 |
2 |
↓
|
|
|
|
Analogs
-
34319463
-

-
34319464
-

-
34319465
-

-
31545286
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2R,4S,5S)-6-[(2S,3R,4R,5S,6R)-3-amino-4,5,6-trihydroxy-tetrahydropyran-2-yl]oxycyclohexane-1,2,3
(1R,2R,4S,5S)-6-[(2S,3R,4R,5S,6R…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLCB3-1-E |
Phospholipase C-beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
PLCG2-1-E |
Phospholipase C-gamma-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.81 |
-16.34 |
-42.44 |
11 |
11 |
1 |
208 |
328.294 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.81 |
-16.82 |
-12.5 |
10 |
11 |
0 |
206 |
327.286 |
2 |
↓
|
|
|
|
Analogs
-
34319463
-

-
34319464
-

-
34319465
-

-
31545286
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2R,4S,5S)-6-[(2S,3S,4R,5S,6R)-3-amino-4,5,6-trihydroxy-tetrahydropyran-2-yl]oxycyclohexane-1,2,3
(1R,2R,4S,5S)-6-[(2S,3S,4R,5S,6R…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLCB3-1-E |
Phospholipase C-beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
PLCG2-1-E |
Phospholipase C-gamma-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.81 |
-16.28 |
-42.89 |
11 |
11 |
1 |
208 |
328.294 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.81 |
-16.6 |
-14.26 |
10 |
11 |
0 |
206 |
327.286 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.56 |
-2.96 |
-18.01 |
4 |
7 |
0 |
108 |
303.355 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.56 |
-2.14 |
-17.73 |
4 |
7 |
0 |
108 |
303.355 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.99 |
-14.34 |
-125.17 |
12 |
11 |
2 |
204 |
367.399 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.99 |
-15.1 |
-15.77 |
10 |
11 |
0 |
201 |
365.383 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.99 |
-14.67 |
-53.71 |
11 |
11 |
1 |
202 |
366.391 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.92 |
-12.71 |
-124.15 |
12 |
10 |
2 |
198 |
323.346 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.92 |
-13.03 |
-53.31 |
11 |
10 |
1 |
196 |
322.338 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.92 |
-13.02 |
-51.4 |
11 |
10 |
1 |
196 |
322.338 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.62 |
-2.94 |
-314.1 |
11 |
8 |
4 |
136 |
336.477 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.62 |
-5.23 |
-43.21 |
8 |
8 |
1 |
129 |
333.453 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.62 |
-4.22 |
-114.68 |
9 |
8 |
2 |
130 |
334.461 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.08 |
-17.98 |
-99.06 |
13 |
11 |
2 |
215 |
342.345 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.08 |
-18.7 |
-10.97 |
11 |
11 |
0 |
212 |
340.329 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.08 |
-18.32 |
-43.68 |
12 |
11 |
1 |
214 |
341.337 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.16 |
-16.84 |
-187.69 |
14 |
11 |
3 |
212 |
356.396 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.16 |
-18.4 |
-10.03 |
11 |
11 |
0 |
204 |
353.372 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.16 |
-16.88 |
-39.21 |
12 |
11 |
1 |
208 |
354.38 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.32 |
-16.37 |
-195.43 |
15 |
11 |
3 |
218 |
355.412 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.32 |
-17.71 |
-41.48 |
13 |
11 |
1 |
211 |
353.396 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.32 |
-15.81 |
-87.18 |
14 |
11 |
2 |
216 |
354.404 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.21 |
-15.25 |
-293.03 |
16 |
11 |
4 |
219 |
370.447 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.21 |
-15.51 |
-85.11 |
14 |
11 |
2 |
216 |
368.431 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.21 |
-16.9 |
-40.84 |
13 |
11 |
1 |
211 |
367.423 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.55 |
-9.34 |
-291.39 |
14 |
9 |
4 |
179 |
338.449 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.55 |
-10.86 |
-41.61 |
11 |
9 |
1 |
171 |
335.425 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.55 |
-10.54 |
-102.97 |
12 |
9 |
2 |
172 |
336.433 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.11 |
-7.23 |
-292.12 |
13 |
9 |
4 |
168 |
352.476 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.11 |
-8.34 |
-112 |
11 |
9 |
2 |
161 |
350.46 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.11 |
-8.73 |
-42.68 |
10 |
9 |
1 |
160 |
349.452 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.11 |
-6.83 |
-287.83 |
13 |
9 |
4 |
168 |
352.476 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.11 |
-8.23 |
-108.2 |
11 |
9 |
2 |
161 |
350.46 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.11 |
-8.63 |
-41.73 |
10 |
9 |
1 |
160 |
349.452 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.11 |
-6.7 |
-293.92 |
13 |
9 |
4 |
168 |
352.476 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.11 |
-8.31 |
-106.41 |
11 |
9 |
2 |
161 |
350.46 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.11 |
-8.62 |
-43.47 |
10 |
9 |
1 |
160 |
349.452 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.77 |
-6.32 |
-264.88 |
14 |
12 |
4 |
209 |
436.554 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.83 |
-6.87 |
-88.1 |
12 |
12 |
2 |
208 |
434.538 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.77 |
-6.9 |
-88.86 |
12 |
12 |
2 |
206 |
434.538 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.81 |
-15.4 |
-102.92 |
12 |
10 |
2 |
195 |
326.346 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.81 |
-16.38 |
-10.19 |
10 |
10 |
0 |
192 |
324.33 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.81 |
-15.78 |
-44.28 |
11 |
10 |
1 |
194 |
325.338 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.82 |
-14.46 |
-189.01 |
13 |
10 |
3 |
191 |
340.397 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.82 |
-14.51 |
-36.38 |
11 |
10 |
1 |
188 |
338.381 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.82 |
-15.67 |
-44.54 |
11 |
10 |
1 |
185 |
338.381 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.04 |
-13.99 |
-197.29 |
14 |
10 |
3 |
197 |
339.413 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.04 |
-13.4 |
-86.8 |
13 |
10 |
2 |
196 |
338.405 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.04 |
-14.82 |
-125.05 |
13 |
10 |
2 |
193 |
338.405 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.20 |
-7.7 |
-306.61 |
13 |
8 |
4 |
158 |
308.423 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.20 |
-7.22 |
-86.53 |
11 |
8 |
2 |
155 |
306.407 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.20 |
-8.84 |
-126.88 |
11 |
8 |
2 |
152 |
306.407 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.61 |
-5.34 |
-307.94 |
12 |
8 |
4 |
147 |
322.45 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.61 |
-5.82 |
-43.6 |
9 |
8 |
1 |
140 |
319.426 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.61 |
-6.5 |
-125.23 |
10 |
8 |
2 |
141 |
320.434 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.61 |
-4.96 |
-302.85 |
12 |
8 |
4 |
147 |
322.45 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.61 |
-5.78 |
-42.83 |
9 |
8 |
1 |
140 |
319.426 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.61 |
-6.54 |
-124.6 |
10 |
8 |
2 |
141 |
320.434 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.89 |
-2.94 |
-215.65 |
10 |
11 |
3 |
169 |
420.531 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.89 |
-3.35 |
-134.53 |
9 |
11 |
2 |
168 |
419.523 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.89 |
-2.06 |
-103.63 |
9 |
11 |
2 |
168 |
419.523 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.00 |
-5 |
-216.14 |
12 |
12 |
3 |
195 |
435.546 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.00 |
-5.41 |
-134.93 |
11 |
12 |
2 |
194 |
434.538 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.00 |
-4.12 |
-103.97 |
11 |
12 |
2 |
194 |
434.538 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.67 |
-1.6 |
-301.24 |
12 |
11 |
4 |
178 |
420.555 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.79 |
-1.01 |
-91.85 |
10 |
11 |
2 |
177 |
418.539 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.79 |
-2 |
-207.03 |
11 |
11 |
3 |
179 |
419.547 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.62 |
-3.65 |
-309.99 |
11 |
8 |
4 |
136 |
336.477 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.62 |
-4.67 |
-37.85 |
8 |
8 |
1 |
129 |
333.453 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.62 |
-5.04 |
-117.21 |
9 |
8 |
2 |
130 |
334.461 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.89 |
-4.63 |
-216.62 |
10 |
11 |
3 |
169 |
420.531 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.89 |
-4.93 |
-137.28 |
9 |
11 |
2 |
168 |
419.523 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.89 |
-3.99 |
-105.08 |
9 |
11 |
2 |
168 |
419.523 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.67 |
-3.29 |
-279.26 |
12 |
11 |
4 |
178 |
420.555 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.79 |
-3.58 |
-182.13 |
11 |
11 |
3 |
179 |
419.547 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.79 |
-2.63 |
-158.58 |
11 |
11 |
3 |
179 |
419.547 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.60 |
-2.09 |
-314.55 |
11 |
8 |
4 |
136 |
350.504 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.60 |
-4.38 |
-42.18 |
8 |
8 |
1 |
129 |
347.48 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.60 |
-3.36 |
-113.82 |
9 |
8 |
2 |
130 |
348.488 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.21 |
-2.31 |
-311.56 |
12 |
10 |
4 |
168 |
407.556 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.21 |
-1.86 |
-98.37 |
10 |
10 |
2 |
165 |
405.54 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.21 |
-2.14 |
-46.64 |
9 |
10 |
1 |
163 |
404.532 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.87 |
-2.08 |
-215.72 |
10 |
11 |
3 |
169 |
434.558 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.87 |
-2.49 |
-133.6 |
9 |
11 |
2 |
168 |
433.55 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.87 |
-1.22 |
-103.31 |
9 |
11 |
2 |
168 |
433.55 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.02 |
-18.24 |
-26.56 |
11 |
15 |
0 |
259 |
516.522 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-amino-N-[(1S,2R,3R,4S,5S,6R)-4-amino-3-[(2S,6S)-6-[(1S)-1-aminoethyl]tetrahydropyran-2-yl]oxy-2,5-
2-amino-N-[(1S,2R,3R,4S,5S,6R)-4…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.10 |
-4.67 |
-195.36 |
11 |
10 |
3 |
171 |
393.505 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.10 |
-5.13 |
-44.63 |
9 |
10 |
1 |
168 |
391.489 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.10 |
-4.73 |
-117.93 |
10 |
10 |
2 |
170 |
392.497 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-amino-N-[(1S,2R,3R,4S,5S,6R)-4-amino-3-[(2S,6R)-6-[(1S)-1-aminoethyl]tetrahydropyran-2-yl]oxy-2,5-
2-amino-N-[(1S,2R,3R,4S,5S,6R)-4…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.10 |
-3.73 |
-175.68 |
11 |
10 |
3 |
171 |
393.505 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.10 |
-4.93 |
-42.74 |
9 |
10 |
1 |
168 |
391.489 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.10 |
-4.58 |
-117.03 |
10 |
10 |
2 |
170 |
392.497 |
6 |
↓
|
|
|
|
|