|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.99 |
-5.96 |
-22.52 |
6 |
11 |
0 |
169 |
339.308 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.99 |
-6.98 |
-24.04 |
6 |
11 |
0 |
169 |
339.308 |
5 |
↓
|
|
|
|
Analogs
-
26578663
-
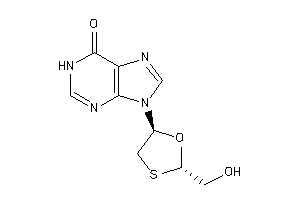
-
5893431
-
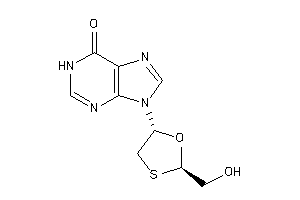
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
1.24 |
-22.97 |
2 |
7 |
0 |
93 |
254.271 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.52 |
0.14 |
-47.69 |
1 |
7 |
-1 |
96 |
253.263 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50527-1-O |
Human Herpesvirus 3 (cluster #1 Of 3), Other |
Other |
720 |
0.23 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
5700 |
0.19 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
11.04 |
-26 |
4 |
13 |
0 |
173 |
542.489 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.60 |
1.52 |
-29.06 |
4 |
7 |
0 |
110 |
283.288 |
2 |
↓
|
|
|
|
Analogs
-
32107391
-
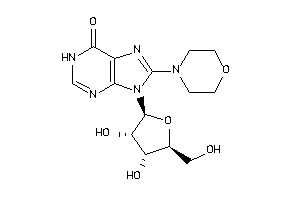
-
4257481
-
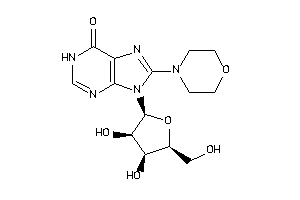
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.55 |
-4.39 |
-32.37 |
4 |
11 |
0 |
146 |
353.335 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.09 |
-5.52 |
-68.87 |
3 |
11 |
-1 |
149 |
352.327 |
3 |
↓
|
|
|
|
Analogs
-
32107391
-

-
4257481
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.55 |
-5.39 |
-32 |
4 |
11 |
0 |
146 |
353.335 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.09 |
-6.51 |
-65.99 |
3 |
11 |
-1 |
149 |
352.327 |
3 |
↓
|
|
|
|
Analogs
-
36152047
-
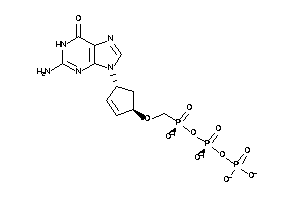
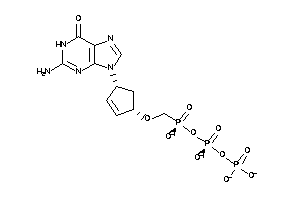
-
13558740
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
300 |
0.30 |
Binding ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
300 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.53 |
2.68 |
-357.56 |
3 |
16 |
-4 |
261 |
483.163 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.53 |
1.52 |
-222.45 |
4 |
16 |
-3 |
258 |
484.171 |
8 |
↓
|
|
|
|
Analogs
-
36152040
-
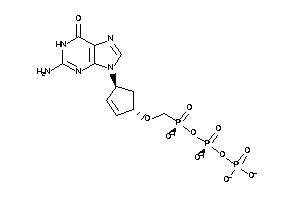
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
300 |
0.30 |
Binding ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
300 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.53 |
2.66 |
-370.92 |
3 |
16 |
-4 |
261 |
483.163 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.53 |
1.5 |
-232.79 |
4 |
16 |
-3 |
258 |
484.171 |
8 |
↓
|
|
|
|
Analogs
-
13642565
-
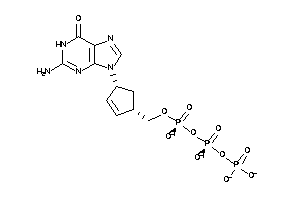
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
37 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.16 |
4.31 |
-369.44 |
3 |
16 |
-4 |
261 |
483.163 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.16 |
3.15 |
-231.72 |
4 |
16 |
-3 |
258 |
484.171 |
8 |
↓
|
|
|
|
Analogs
-
31517421
-
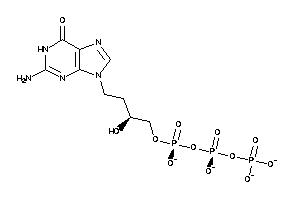
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.82 |
2.22 |
-374.74 |
3 |
17 |
-4 |
270 |
487.151 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.82 |
1.07 |
-236.34 |
4 |
17 |
-3 |
267 |
488.159 |
8 |
↓
|
|
|
|
Analogs
-
31517421
-

-
25760528
-
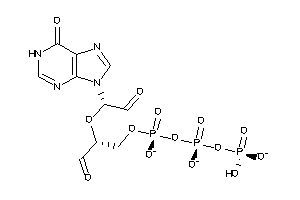
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.82 |
2.2 |
-375.8 |
3 |
17 |
-4 |
270 |
487.151 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.82 |
1.05 |
-237.01 |
4 |
17 |
-3 |
267 |
488.159 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.88 |
-1 |
-157.92 |
3 |
12 |
-2 |
186 |
390.22 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.42 |
-2.89 |
-212.88 |
2 |
12 |
-3 |
189 |
389.212 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.88 |
-2.16 |
-65.3 |
4 |
12 |
-1 |
183 |
391.228 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.88 |
-1.66 |
-152.11 |
3 |
12 |
-2 |
186 |
390.22 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.42 |
-3.56 |
-210.84 |
2 |
12 |
-3 |
189 |
389.212 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.88 |
-2.81 |
-60.84 |
4 |
12 |
-1 |
183 |
391.228 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.88 |
-2.73 |
-152.48 |
3 |
12 |
-2 |
186 |
390.22 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.42 |
-4.62 |
-208.56 |
2 |
12 |
-3 |
189 |
389.212 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.88 |
-3.89 |
-60.37 |
4 |
12 |
-1 |
183 |
391.228 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.88 |
-2.56 |
-155.78 |
3 |
12 |
-2 |
186 |
390.22 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.42 |
-4.46 |
-212.14 |
2 |
12 |
-3 |
189 |
389.212 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.88 |
-3.72 |
-62.41 |
4 |
12 |
-1 |
183 |
391.228 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.81 |
-0.95 |
-159.99 |
3 |
12 |
-2 |
186 |
394.664 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.35 |
-3.35 |
-213.49 |
2 |
12 |
-3 |
189 |
393.656 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.81 |
-2.1 |
-66.74 |
4 |
12 |
-1 |
183 |
395.672 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.81 |
-1.62 |
-156.49 |
3 |
12 |
-2 |
186 |
394.664 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.35 |
-4.03 |
-212.52 |
2 |
12 |
-3 |
189 |
393.656 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.81 |
-2.78 |
-64.39 |
4 |
12 |
-1 |
183 |
395.672 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.81 |
-2.69 |
-155.5 |
3 |
12 |
-2 |
186 |
394.664 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.35 |
-5.08 |
-209.14 |
2 |
12 |
-3 |
189 |
393.656 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.81 |
-3.84 |
-62.78 |
4 |
12 |
-1 |
183 |
395.672 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.81 |
-2.5 |
-158.19 |
3 |
12 |
-2 |
186 |
394.664 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.35 |
-4.9 |
-213.75 |
2 |
12 |
-3 |
189 |
393.656 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.81 |
-3.66 |
-64.44 |
4 |
12 |
-1 |
183 |
395.672 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-1.54 |
-161.44 |
3 |
12 |
-2 |
186 |
378.209 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.66 |
-3.74 |
-214.6 |
2 |
12 |
-3 |
189 |
377.201 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.11 |
-2.7 |
-68.04 |
4 |
12 |
-1 |
183 |
379.217 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-2.21 |
-158 |
3 |
12 |
-2 |
186 |
378.209 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.66 |
-4.42 |
-213.79 |
2 |
12 |
-3 |
189 |
377.201 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.11 |
-3.37 |
-65.89 |
4 |
12 |
-1 |
183 |
379.217 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-3.28 |
-156.84 |
3 |
12 |
-2 |
186 |
378.209 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.66 |
-5.47 |
-210.26 |
2 |
12 |
-3 |
189 |
377.201 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.11 |
-4.43 |
-64.15 |
4 |
12 |
-1 |
183 |
379.217 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-3.09 |
-159.58 |
3 |
12 |
-2 |
186 |
378.209 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.66 |
-5.28 |
-215.01 |
2 |
12 |
-3 |
189 |
377.201 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.11 |
-4.25 |
-65.68 |
4 |
12 |
-1 |
183 |
379.217 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.71 |
-1.12 |
-160.33 |
3 |
12 |
-2 |
186 |
372.23 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.25 |
-3.01 |
-219 |
2 |
12 |
-3 |
189 |
371.222 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.71 |
-2.28 |
-67.11 |
4 |
12 |
-1 |
183 |
373.238 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.71 |
-1.78 |
-155.03 |
3 |
12 |
-2 |
186 |
372.23 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.25 |
-3.68 |
-216.49 |
2 |
12 |
-3 |
189 |
371.222 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.71 |
-2.94 |
-63.18 |
4 |
12 |
-1 |
183 |
373.238 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.71 |
-2.85 |
-155.35 |
3 |
12 |
-2 |
186 |
372.23 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.25 |
-4.74 |
-214.63 |
2 |
12 |
-3 |
189 |
371.222 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.71 |
-4 |
-62.58 |
4 |
12 |
-1 |
183 |
373.238 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.71 |
-2.68 |
-158.43 |
3 |
12 |
-2 |
186 |
372.23 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.25 |
-4.57 |
-218.3 |
2 |
12 |
-3 |
189 |
371.222 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.71 |
-3.83 |
-64.36 |
4 |
12 |
-1 |
183 |
373.238 |
5 |
↓
|
|
|
|
Analogs
-
27888045
-
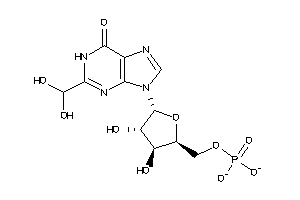
-
27888052
-
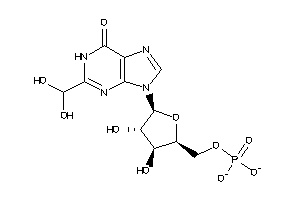
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.70 |
-6.97 |
-158.36 |
5 |
14 |
-2 |
226 |
392.217 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.25 |
-8.98 |
-216.88 |
4 |
14 |
-3 |
229 |
391.209 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.70 |
-8.12 |
-65.93 |
6 |
14 |
-1 |
223 |
393.225 |
5 |
↓
|
|
|
|
Analogs
-
27888045
-

-
27888052
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.70 |
-7.64 |
-155.39 |
5 |
14 |
-2 |
226 |
392.217 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.25 |
-9.66 |
-214.67 |
4 |
14 |
-3 |
229 |
391.209 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.70 |
-8.79 |
-62.79 |
6 |
14 |
-1 |
223 |
393.225 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYPH-1-E |
Thymidine Phosphorylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.17 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYPH_HUMAN |
P19971
|
Thymidine Phosphorylase, Human |
4400 |
0.17 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.94 |
13.95 |
-24.31 |
2 |
9 |
0 |
112 |
606.723 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYPH-2-E |
Thymidine Phosphorylase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.19 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYPH_HUMAN |
P19971
|
Thymidine Phosphorylase, Human |
4500 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
8.91 |
-23.8 |
3 |
9 |
0 |
123 |
544.995 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.77 |
6.84 |
-58.75 |
2 |
9 |
-1 |
126 |
543.987 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYPH-1-E |
Thymidine Phosphorylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.19 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYPH_HUMAN |
P19971
|
Thymidine Phosphorylase, Human |
2300 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.52 |
12.77 |
-24.34 |
2 |
9 |
0 |
112 |
564.642 |
9 |
↓
|
|
|
|
Analogs
-
44699093
-
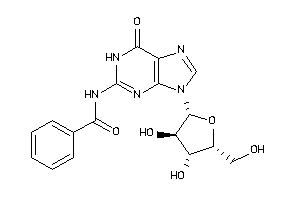
-
4293804
-
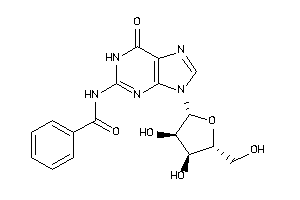
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
-1.67 |
-24.61 |
5 |
11 |
0 |
163 |
387.352 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.27 |
-3.5 |
-73.41 |
4 |
11 |
-1 |
166 |
386.344 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
20192528
-
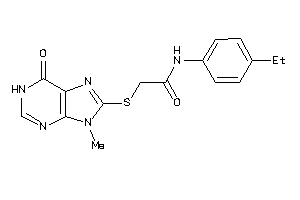
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
-2 |
-20.82 |
2 |
7 |
0 |
93 |
357.439 |
5 |
↓
|
|
|
|
Analogs
-
12655466
-
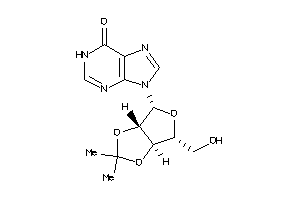
-
13546310
-
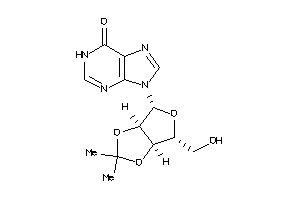
-
1319989
-
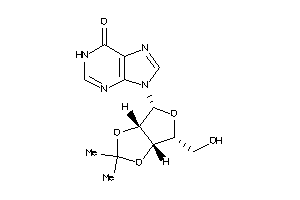
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.89 |
0.17 |
-23.85 |
2 |
9 |
0 |
112 |
308.294 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.43 |
-1.89 |
-60.72 |
1 |
9 |
-1 |
115 |
307.286 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
8382313
-
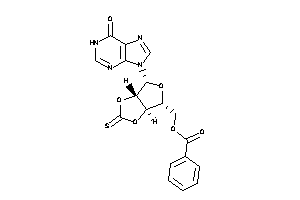
-
8382314
-
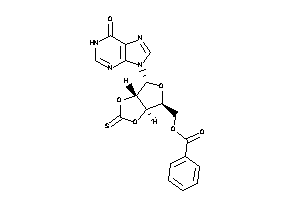
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3aS,4S,6R,6aS)-6-(6-hydroxypurin-9-yl)-2-thioxo-3a,4,6,6a-tetrahydrofuro[4,3-d][1,3]dioxol-4-yl]me
[(3aS,4S,6R,6aS)-6-(6-hydroxypur…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
7.88 |
-35.19 |
1 |
10 |
0 |
118 |
414.399 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.61 |
6.79 |
-62.27 |
0 |
10 |
-1 |
121 |
413.391 |
5 |
↓
|
|
|
|
Analogs
-
8382313
-

-
8382314
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3aS,4S,6S,6aS)-6-(6-hydroxypurin-9-yl)-2-thioxo-3a,4,6,6a-tetrahydrofuro[4,3-d][1,3]dioxol-4-yl]me
[(3aS,4S,6S,6aS)-6-(6-hydroxypur…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
7.78 |
-25.15 |
1 |
10 |
0 |
118 |
414.399 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.61 |
5.72 |
-54.69 |
0 |
10 |
-1 |
121 |
413.391 |
5 |
↓
|
|
|
|
Analogs
-
8382313
-

-
8382314
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3aS,4R,6R,6aS)-6-(6-hydroxypurin-9-yl)-2-thioxo-3a,4,6,6a-tetrahydrofuro[4,3-d][1,3]dioxol-4-yl]me
[(3aS,4R,6R,6aS)-6-(6-hydroxypur…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
8.37 |
-41.4 |
1 |
10 |
0 |
118 |
414.399 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.61 |
7.27 |
-63.18 |
0 |
10 |
-1 |
121 |
413.391 |
5 |
↓
|
|
|
|
Analogs
-
8382313
-

-
8382314
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3aS,4R,6S,6aS)-6-(6-hydroxypurin-9-yl)-2-thioxo-3a,4,6,6a-tetrahydrofuro[4,3-d][1,3]dioxol-4-yl]me
[(3aS,4R,6S,6aS)-6-(6-hydroxypur…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
7.93 |
-30.95 |
1 |
10 |
0 |
118 |
414.399 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.61 |
5.87 |
-64.74 |
0 |
10 |
-1 |
121 |
413.391 |
5 |
↓
|
|
|
|
Analogs
-
8738349
-
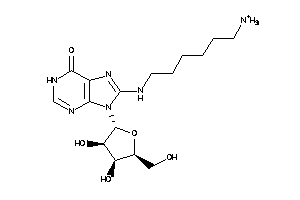
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.29 |
-3.77 |
-65.16 |
8 |
11 |
1 |
173 |
383.429 |
9 |
↓
|
|
|
|
Analogs
-
8738349
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.29 |
-3.92 |
-63.75 |
8 |
11 |
1 |
173 |
383.429 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.81 |
-4.18 |
-39.67 |
9 |
14 |
1 |
215 |
443.4 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.81 |
-4.41 |
-39.72 |
9 |
14 |
1 |
215 |
443.4 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.03 |
1.88 |
-25.28 |
2 |
6 |
0 |
84 |
234.259 |
2 |
↓
|
|