|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
8.22 |
-37.47 |
1 |
3 |
1 |
20 |
316.425 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.27 |
6.81 |
-27.22 |
1 |
3 |
1 |
20 |
316.425 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.27 |
5.84 |
-4.55 |
0 |
3 |
0 |
19 |
315.417 |
1 |
↓
|
|
|
|
Analogs
-
4214594
-
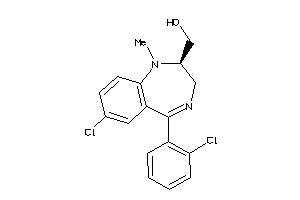
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
-3.62 |
-7.29 |
1 |
3 |
0 |
35 |
335.234 |
2 |
↓
|
|
|
|
Analogs
-
5666944
-
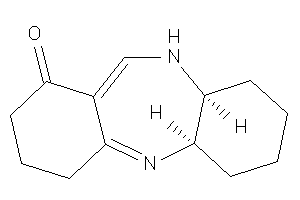
-
5666964
-
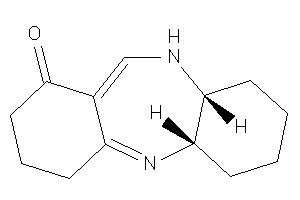
-
5666968
-
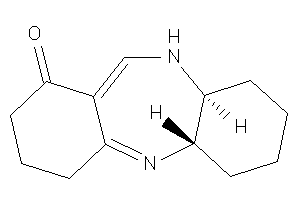
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aR,11aR)-1,2,3,4,4a,5,8,9,10,11a-decahydrobenzo[b][1,4]benzodiazepin-7-one
(4aR,11aR)-1,2,3,4,4a,5,8,9,10,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.48 |
4.24 |
-7.43 |
1 |
3 |
0 |
41 |
218.3 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.81 |
-3.39 |
-99.36 |
2 |
3 |
2 |
45 |
220.316 |
0 |
↓
|
|
|
|
Analogs
-
36216017
-
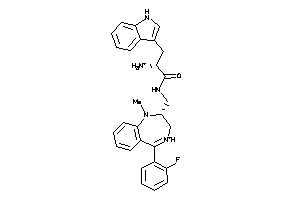
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
9.7 |
-90.09 |
6 |
6 |
2 |
90 |
471.58 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.79 |
8.8 |
-13.53 |
4 |
6 |
0 |
87 |
469.564 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.79 |
9.09 |
-52.57 |
5 |
6 |
1 |
88 |
470.572 |
6 |
↓
|
|
|
|
Analogs
-
36216014
-
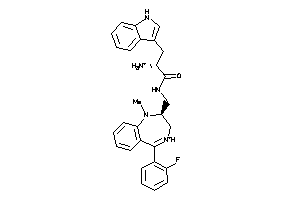
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
9.67 |
-89.71 |
6 |
6 |
2 |
90 |
471.58 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.79 |
8.79 |
-13.25 |
4 |
6 |
0 |
87 |
469.564 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.79 |
9.08 |
-52.15 |
5 |
6 |
1 |
88 |
470.572 |
6 |
↓
|
|
|
|
Analogs
-
36216027
-
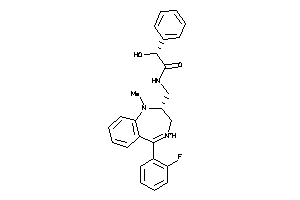
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.28 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR_RAT |
P30551
|
Cholecystokinin A Receptor, Rat |
600 |
0.28 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 1μM
|
|
CCKAR_RAT |
P30551
|
Cholecystokinin A Receptor, Rat |
600 |
0.28 |
Binding ≤ 10μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
9.63 |
-35.47 |
3 |
5 |
1 |
67 |
418.492 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.79 |
9.02 |
-13.48 |
2 |
5 |
0 |
65 |
417.484 |
5 |
↓
|
|
|
|
Analogs
-
36216024
-
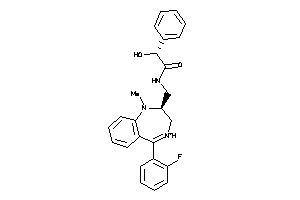
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR-1-E |
Cholecystokinin A Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.28 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCKAR_RAT |
P30551
|
Cholecystokinin A Receptor, Rat |
600 |
0.28 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 1μM
|
|
CCKAR_RAT |
P30551
|
Cholecystokinin A Receptor, Rat |
600 |
0.28 |
Binding ≤ 10μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
7 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
9.61 |
-35.66 |
3 |
5 |
1 |
67 |
418.492 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.79 |
9.02 |
-13.44 |
2 |
5 |
0 |
65 |
417.484 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.97 |
18.05 |
-74.31 |
1 |
6 |
0 |
66 |
507.078 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.97 |
15.83 |
-54.38 |
0 |
6 |
-1 |
64 |
506.07 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
17.32 |
-69.82 |
1 |
6 |
0 |
66 |
472.633 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.56 |
15.11 |
-53.92 |
0 |
6 |
-1 |
64 |
471.625 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.56 |
17.81 |
-134.59 |
2 |
6 |
1 |
67 |
473.641 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.21 |
14.55 |
-13.58 |
0 |
8 |
0 |
77 |
516.642 |
6 |
↓
|
|