|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.89 |
4.75 |
-73.51 |
8 |
11 |
1 |
183 |
460.559 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.89 |
3.62 |
-89.23 |
7 |
11 |
0 |
178 |
459.551 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
3.32 |
-6.29 |
2 |
4 |
0 |
50 |
224.235 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
3.6 |
-5.55 |
2 |
4 |
0 |
50 |
224.235 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
3.59 |
-4.91 |
2 |
4 |
0 |
50 |
224.235 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
3.32 |
-5.66 |
2 |
4 |
0 |
50 |
224.235 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
1.02 |
-3.01 |
4 |
3 |
0 |
50 |
163.224 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.01 |
0.9 |
-42.3 |
5 |
3 |
1 |
52 |
164.232 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.01 |
1.58 |
-39.89 |
5 |
3 |
1 |
55 |
164.232 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
0.83 |
-3.13 |
4 |
3 |
0 |
50 |
163.224 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.01 |
1.39 |
-38.51 |
5 |
3 |
1 |
55 |
164.232 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.01 |
0.7 |
-42.73 |
5 |
3 |
1 |
52 |
164.232 |
1 |
↓
|
|
|
|
Analogs
-
39237872
-
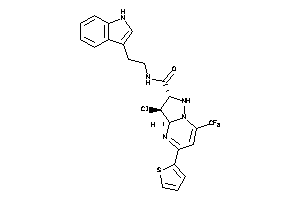
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
5.47 |
-42.11 |
4 |
7 |
1 |
87 |
445.878 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.50 |
5.01 |
-14.49 |
3 |
7 |
0 |
85 |
444.87 |
6 |
↓
|
|
|
|
Analogs
-
39237872
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
5.73 |
-43.51 |
4 |
7 |
1 |
87 |
445.878 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.50 |
5.26 |
-16.19 |
3 |
7 |
0 |
85 |
444.87 |
6 |
↓
|
|
|
|
Analogs
-
39237872
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
5.85 |
-45.17 |
4 |
7 |
1 |
87 |
445.878 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.50 |
5.38 |
-17.22 |
3 |
7 |
0 |
85 |
444.87 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
7.05 |
-12.31 |
5 |
8 |
0 |
103 |
336.403 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.28 |
7.58 |
-40.28 |
6 |
8 |
1 |
104 |
337.411 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.28 |
8.37 |
-168.44 |
8 |
8 |
3 |
110 |
339.427 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
6.82 |
-10.48 |
5 |
8 |
0 |
103 |
336.403 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.28 |
7.38 |
-39.32 |
6 |
8 |
1 |
104 |
337.411 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.28 |
8.37 |
-166.64 |
8 |
8 |
3 |
110 |
339.427 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
6.72 |
-12.05 |
5 |
8 |
0 |
103 |
336.403 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.28 |
7.25 |
-40.54 |
6 |
8 |
1 |
104 |
337.411 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.28 |
8.35 |
-167.98 |
8 |
8 |
3 |
110 |
339.427 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
6.68 |
-12.03 |
5 |
8 |
0 |
103 |
336.403 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.28 |
7.22 |
-41.34 |
6 |
8 |
1 |
104 |
337.411 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.28 |
8.31 |
-166.51 |
8 |
8 |
3 |
110 |
339.427 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB_HUMAN |
P07858
|
Cathepsin B, Human |
119 |
0.26 |
Binding ≤ 1μM
|
|
CATB_RAT |
P00787
|
Cathepsin B, Rat |
119 |
0.26 |
Binding ≤ 1μM
|
|
CATK_HUMAN |
P43235
|
Cathepsin K, Human |
3.5 |
0.32 |
Binding ≤ 1μM
|
|
CATL1_HUMAN |
P07711
|
Cathepsin L, Human |
370 |
0.24 |
Binding ≤ 1μM
|
|
CATL3_PARTE |
Q94715
|
Cathepsin L2, Parte |
370 |
0.24 |
Binding ≤ 1μM
|
|
CATS_HUMAN |
P25774
|
Cathepsin S, Human |
434 |
0.24 |
Binding ≤ 1μM
|
|
CYSP_TRYCR |
P25779
|
Cruzipain, Trycr |
93.2 |
0.27 |
Binding ≤ 1μM
|
|
LMCPB_LEIME |
P36400
|
Cysteine Proteinase B, Leime |
49.5 |
0.28 |
Binding ≤ 1μM
|
|
CATB_RAT |
P00787
|
Cathepsin B, Rat |
119 |
0.26 |
Binding ≤ 10μM
|
|
CATB_HUMAN |
P07858
|
Cathepsin B, Human |
119 |
0.26 |
Binding ≤ 10μM
|
|
CATK_HUMAN |
P43235
|
Cathepsin K, Human |
3.5 |
0.32 |
Binding ≤ 10μM
|
|
CATL1_HUMAN |
P07711
|
Cathepsin L, Human |
370 |
0.24 |
Binding ≤ 10μM
|
|
CATL3_PARTE |
Q94715
|
Cathepsin L2, Parte |
370 |
0.24 |
Binding ≤ 10μM
|
|
CATS_HUMAN |
P25774
|
Cathepsin S, Human |
434 |
0.24 |
Binding ≤ 10μM
|
|
CYSP_TRYCR |
P25779
|
Cruzipain, Trycr |
93.2 |
0.27 |
Binding ≤ 10μM
|
|
LMCPB_LEIME |
P36400
|
Cysteine Proteinase B, Leime |
49.5 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
10.33 |
-13.05 |
2 |
8 |
0 |
99 |
504.631 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.13 |
-1.61 |
-11.16 |
1 |
5 |
0 |
64 |
168.152 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
4131144
-
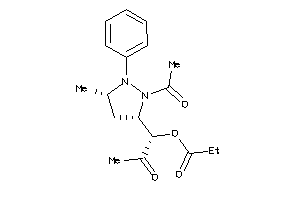
-
4131145
-
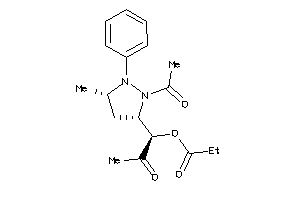
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
9.52 |
-13.33 |
0 |
6 |
0 |
67 |
332.4 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
8.18 |
-6.38 |
1 |
4 |
0 |
36 |
309.413 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
8.26 |
-6.31 |
1 |
4 |
0 |
36 |
309.413 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
7.73 |
-9.37 |
1 |
4 |
0 |
36 |
309.413 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
8.33 |
-8.92 |
1 |
4 |
0 |
36 |
309.413 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
7.92 |
-8.67 |
1 |
4 |
0 |
36 |
295.386 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
7.67 |
-6.24 |
1 |
4 |
0 |
36 |
295.386 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
4.65 |
-9.48 |
0 |
6 |
0 |
59 |
270.358 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
6.27 |
-22.71 |
0 |
7 |
0 |
84 |
368.455 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.36 |
2.81 |
-11.65 |
2 |
6 |
0 |
65 |
348.472 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.36 |
4.3 |
-41.65 |
3 |
6 |
1 |
66 |
349.48 |
5 |
↓
|
|